In this problem set, you will implement the sparse autoencoder algorithm, and show how it discovers that edges are a good representation for natural images. (Images provided by Bruno Olshausen.) The sparse autoencoder algorithm is described in the lecture notes found on the course website.
In the file sparseae_exercise.zip, we have provided some starter code in Matlab. You should write your code at the places indicated in the files ("YOUR CODE HERE"). You have to complete the following files: sampleIMAGES.m, sparseAutoencoderCost.m, computeNumericalGradient.m. The starter code in train.m shows how these functions are used.
Specifically, in this exercise you will implement a sparse autoencoder, trained with 8×8 image patches using the L-BFGS optimization algorithm.
A note on the software: The provided .zip file includes a subdirectory minFunc with 3rd party software implementing L-BFGS, that is licensed under a Creative Commons, Attribute, Non-Commercial license. If you need to use this software for commercial purposes, you can download and use a different function (fminlbfgs) that can serve the same purpose, but runs ~3x slower for this exercise (and thus is less recommended). You can read more about this in the Fminlbfgs_Details page.
- %% CS294A/CS294W Programming Assignment Starter Code
-
- % Instructions
- % ------------
- %
- % This file contains code that helps you get started on the
- % programming assignment. You will need to complete the code in sampleIMAGES.m,
- % sparseAutoencoderCost.m and computeNumericalGradient.m.
- % For the purpose of completing the assignment, you do not need to
- % change the code in this file.
- %
- %%======================================================================
- %% STEP 0: Here we provide the relevant parameters values that will
- % allow your sparse autoencoder to get good filters; you do not need to
- % change the parameters below.
-
- visibleSize = 8*8; % number of input units
- hiddenSize = 25; % number of hidden units
- sparsityParam = 0.01; % desired average activation of the hidden units.
- % (This was denoted by the Greek alphabet rho, which looks like a lower-case "p",
- % in the lecture notes).
- lambda = 0.0001; % weight decay parameter
- beta = 3; % weight of sparsity penalty term
-
- %%======================================================================
- %% STEP 1: Implement sampleIMAGES
- %
- % After implementing sampleIMAGES, the display_network command should
- % display a random sample of 200 patches from the dataset
-
- patches = sampleIMAGES;
- display_network(patches(:,randi(size(patches,2),200,1)),8);
-
-
- % Obtain random parameters theta
- theta = initializeParameters(hiddenSize, visibleSize);
-
- %%======================================================================
- %% STEP 2: Implement sparseAutoencoderCost
- %
- % You can implement all of the components (squared error cost, weight decay term,
- % sparsity penalty) in the cost function at once, but it may be easier to do
- % it step-by-step and run gradient checking (see STEP 3) after each step. We
- % suggest implementing the sparseAutoencoderCost function using the following steps:
- %
- % (a) Implement forward propagation in your neural network, and implement the
- % squared error term of the cost function. Implement backpropagation to
- % compute the derivatives. Then (using lambda=beta=0), run Gradient Checking
- % to verify that the calculations corresponding to the squared error cost
- % term are correct.
- %
- % (b) Add in the weight decay term (in both the cost function and the derivative
- % calculations), then re-run Gradient Checking to verify correctness.
- %
- % (c) Add in the sparsity penalty term, then re-run Gradient Checking to
- % verify correctness.
- %
- % Feel free to change the training settings when debugging your
- % code. (For example, reducing the training set size or
- % number of hidden units may make your code run faster; and setting beta
- % and/or lambda to zero may be helpful for debugging.) However, in your
- % final submission of the visualized weights, please use parameters we
- % gave in Step 0 above.
-
- [cost, grad] = sparseAutoencoderCost(theta, visibleSize, hiddenSize, lambda, ...
- sparsityParam, beta, patches);
-
- %%======================================================================
- %% STEP 3: Gradient Checking
- %
- % Hint: If you are debugging your code, performing gradient checking on smaller models
- % and smaller training sets (e.g., using only 10 training examples and 1-2 hidden
- % units) may speed things up.
-
- % First, lets make sure your numerical gradient computation is correct for a
- % simple function. After you have implemented computeNumericalGradient.m,
- % run the following:
- checkNumericalGradient();
-
- % Now we can use it to check your cost function and derivative calculations
- % for the sparse autoencoder.
- numgrad = computeNumericalGradient( @(x) sparseAutoencoderCost(x, visibleSize, ...
- hiddenSize, lambda, ...
- sparsityParam, beta, ...
- patches), theta);
-
- % Use this to visually compare the gradients side by side
- disp([numgrad grad]);
-
- % Compare numerically computed gradients with the ones obtained from backpropagation
- diff = norm(numgrad-grad)/norm(numgrad+grad);
- disp(diff); % Should be small. In our implementation, these values are
- % usually less than 1e-9.
-
- % When you got this working, Congratulations!!!
-
- %%======================================================================
- %% STEP 4: After verifying that your implementation of
- % sparseAutoencoderCost is correct, You can start training your sparse
- % autoencoder with minFunc (L-BFGS).
-
- % Randomly initialize the parameters
- theta = initializeParameters(hiddenSize, visibleSize);
-
- % Use minFunc to minimize the function
- addpath minFunc/
- options.Method = 'lbfgs'; % Here, we use L-BFGS to optimize our cost
- % function. Generally, for minFunc to work, you
- % need a function pointer with two outputs: the
- % function value and the gradient. In our problem,
- % sparseAutoencoderCost.m satisfies this.
- options.maxIter = 400; % Maximum number of iterations of L-BFGS to run
- options.display = 'on';
-
-
- [opttheta, cost] = minFunc( @(p) sparseAutoencoderCost(p, ...
- visibleSize, hiddenSize, ...
- lambda, sparsityParam, ...
- beta, patches), ...
- theta, options);
-
- %%======================================================================
- %% STEP 5: Visualization
-
- W1 = reshape(opttheta(1:hiddenSize*visibleSize), hiddenSize, visibleSize);
- display_network(W1', 12);
-
- print -djpeg weights.jpg % save the visualization to a file
- %% CS294A/CS294W Programming Assignment Starter Code
- % Instructions
- % ------------
- %
- % This file contains code that helps you get started on the
- % programming assignment. You will need to complete the code in sampleIMAGES.m,
- % sparseAutoencoderCost.m and computeNumericalGradient.m.
- % For the purpose of completing the assignment, you do not need to
- % change the code in this file.
- %
- %%======================================================================
- %% STEP 0: Here we provide the relevant parameters values that will
- % allow your sparse autoencoder to get good filters; you do not need to
- % change the parameters below.
- visibleSize = 8*8; % number of input units
- hiddenSize = 25; % number of hidden units
- sparsityParam = 0.01; % desired average activation of the hidden units.
- % (This was denoted by the Greek alphabet rho, which looks like a lower-case "p",
- % in the lecture notes).
- lambda = 0.0001; % weight decay parameter
- beta = 3; % weight of sparsity penalty term
- %%======================================================================
- %% STEP 1: Implement sampleIMAGES
- %
- % After implementing sampleIMAGES, the display_network command should
- % display a random sample of 200 patches from the dataset
- patches = sampleIMAGES;
- display_network(patches(:,randi(size(patches,2),200,1)),8);
- % Obtain random parameters theta
- theta = initializeParameters(hiddenSize, visibleSize);
- %%======================================================================
- %% STEP 2: Implement sparseAutoencoderCost
- %
- % You can implement all of the components (squared error cost, weight decay term,
- % sparsity penalty) in the cost function at once, but it may be easier to do
- % it step-by-step and run gradient checking (see STEP 3) after each step. We
- % suggest implementing the sparseAutoencoderCost function using the following steps:
- %
- % (a) Implement forward propagation in your neural network, and implement the
- % squared error term of the cost function. Implement backpropagation to
- % compute the derivatives. Then (using lambda=beta=0), run Gradient Checking
- % to verify that the calculations corresponding to the squared error cost
- % term are correct.
- %
- % (b) Add in the weight decay term (in both the cost function and the derivative
- % calculations), then re-run Gradient Checking to verify correctness.
- %
- % (c) Add in the sparsity penalty term, then re-run Gradient Checking to
- % verify correctness.
- %
- % Feel free to change the training settings when debugging your
- % code. (For example, reducing the training set size or
- % number of hidden units may make your code run faster; and setting beta
- % and/or lambda to zero may be helpful for debugging.) However, in your
- % final submission of the visualized weights, please use parameters we
- % gave in Step 0 above.
- [cost, grad] = sparseAutoencoderCost(theta, visibleSize, hiddenSize, lambda, ...
- sparsityParam, beta, patches);
- %%======================================================================
- %% STEP 3: Gradient Checking
- %
- % Hint: If you are debugging your code, performing gradient checking on smaller models
- % and smaller training sets (e.g., using only 10 training examples and 1-2 hidden
- % units) may speed things up.
- % First, lets make sure your numerical gradient computation is correct for a
- % simple function. After you have implemented computeNumericalGradient.m,
- % run the following:
- checkNumericalGradient();
- % Now we can use it to check your cost function and derivative calculations
- % for the sparse autoencoder.
- numgrad = computeNumericalGradient( @(x) sparseAutoencoderCost(x, visibleSize, ...
- hiddenSize, lambda, ...
- sparsityParam, beta, ...
- patches), theta);
- % Use this to visually compare the gradients side by side
- disp([numgrad grad]);
- % Compare numerically computed gradients with the ones obtained from backpropagation
- diff = norm(numgrad-grad)/norm(numgrad+grad);
- disp(diff); % Should be small. In our implementation, these values are
- % usually less than 1e-9.
- % When you got this working, Congratulations!!!
- %%======================================================================
- %% STEP 4: After verifying that your implementation of
- % sparseAutoencoderCost is correct, You can start training your sparse
- % autoencoder with minFunc (L-BFGS).
- % Randomly initialize the parameters
- theta = initializeParameters(hiddenSize, visibleSize);
- % Use minFunc to minimize the function
- addpath minFunc/
- options.Method = 'lbfgs'; % Here, we use L-BFGS to optimize our cost
- % function. Generally, for minFunc to work, you
- % need a function pointer with two outputs: the
- % function value and the gradient. In our problem,
- % sparseAutoencoderCost.m satisfies this.
- options.maxIter = 400; % Maximum number of iterations of L-BFGS to run
- options.display = 'on';
- [opttheta, cost] = minFunc( @(p) sparseAutoencoderCost(p, ...
- visibleSize, hiddenSize, ...
- lambda, sparsityParam, ...
- beta, patches), ...
- theta, options);
- %%======================================================================
- %% STEP 5: Visualization
- W1 = reshape(opttheta(1:hiddenSize*visibleSize), hiddenSize, visibleSize);
- display_network(W1', 12);
- print -djpeg weights.jpg % save the visualization to a file
Step 1: Generate training set
The first step is to generate a training set. To get a single training example x, randomly pick one of the 10 images, then randomly sample an 8×8 image patch from the selected image, and convert the image patch (either in row-major order or column-major order; it doesn't matter) into a 64-dimensional vector to get a training example 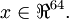
Complete the code in sampleIMAGES.m. Your code should sample 10000 image patches and concatenate them into a 64×10000 matrix.
To make sure your implementation is working, run the code in "Step 1" of train.m. This should result in a plot of a random sample of 200 patches from the dataset.
sampleIMAGES.m的代码:
- function patches = sampleIMAGES()
- % sampleIMAGES
- % Returns 10000 patches for training
- load IMAGES; % load images from disk
- patchsize = 8; % we'll use 8x8 patches
- numpatches = 10000;
- % Initialize patches with zeros. Your code will fill in this matrix--one
- % column per patch, 10000 columns.
- patches = zeros(patchsize*patchsize, numpatches);
- %% ---------- YOUR CODE HERE --------------------------------------
- % Instructions: Fill in the variable called "patches" using data
- % from IMAGES.
- %
- % IMAGES is a 3D array containing 10 images
- % For instance, IMAGES(:,:,6) is a 512x512 array containing the 6th image,
- % and you can type "imagesc(IMAGES(:,:,6)), colormap gray;" to visualize
- % it. (The contrast on these images look a bit off because they have
- % been preprocessed using using "whitening." See the lecture notes for
- % more details.) As a second example, IMAGES(21:30,21:30,1) is an image
- % patch corresponding to the pixels in the block (21,21) to (30,30) of
- % Image 1
- imageCount=size(IMAGES,3);
- for patchNum = 1:10000
- imageNum=randi(1,imageCount);
- [rowNum colNum] = size(IMAGES(:,:,imageNum));
- xPos = randi([1,rowNum-patchsize+1]);
- yPos = randi([1,colNum-patchsize+1]);
- patches(:,patchNum) = reshape(IMAGES(xPos:xPos+7,yPos:yPos+7,imageNum),64,1); end
- end
- %% ---------------------------------------------------------------
- % For the autoencoder to work well we need to normalize the data
- % Specifically, since the output of the network is bounded between [0,1]
- % (due to the sigmoid activation function), we have to make sure
- % the range of pixel values is also bounded between [0,1]
- patches = normalizeData(patches);
- end
- %% ---------------------------------------------------------------
- function patches = normalizeData(patches)
- % Squash data to [0.1, 0.9] since we use sigmoid as the activation
- % function in the output layer
- % Remove DC (mean of images).
- patches = bsxfun(@minus, patches, mean(patches));
- % Truncate to +/-3 standard deviations and scale to -1 to 1
- pstd = 3 * std(patches(:));
- patches = max(min(patches, pstd), -pstd) / pstd;
- % Rescale from [-1,1] to [0.1,0.9]
- patches = (patches + 1) * 0.4 + 0.1;
- end
Implementational tip: When we run our implemented sampleImages(), it takes under 5 seconds. If your implementation takes over 30 seconds, it may be because you are accidentally making a copy of an entire 512×512 image each time you're picking a random image. By copying a 512×512 image 10000 times, this can make your implementation much less efficient. While this doesn't slow down your code significantly for this exercise (because we have only 10000 examples), when we scale to much larger problems later this quarter with 106 or more examples, this will significantly slow down your code. Please implement sampleIMAGES so that you aren't making a copy of an entire 512×512 image each time you need to cut out an 8x8 image patch.
Step 2: Sparse autoencoder objective
Implement code to compute the sparse autoencoder cost function Jsparse(W,b) (Section 3 of the lecture notes) and the corresponding derivatives of Jsparse with respect to the different parameters. Use the sigmoid function for the activation function, 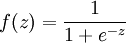 . In particular, complete the code in sparseAutoencoderCost.m.
. In particular, complete the code in sparseAutoencoderCost.m.
The sparse autoencoder is parameterized by matrices 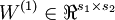 ,
, 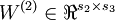 vectors
vectors 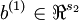 ,
, 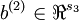 . However, for subsequent notational convenience, we will "unroll" all of these parameters into a very long parameter vector θ with s1s2 + s2s3 +s2 + s3 elements. The code for converting between the (W(1),W(2),b(1),b(2)) and the θ parameterization is already provided in the starter code.
. However, for subsequent notational convenience, we will "unroll" all of these parameters into a very long parameter vector θ with s1s2 + s2s3 +s2 + s3 elements. The code for converting between the (W(1),W(2),b(1),b(2)) and the θ parameterization is already provided in the starter code.
Implementational tip: The objective Jsparse(W,b) contains 3 terms, corresponding to the squared error term, the weight decay term, and the sparsity penalty. You're welcome to implement this however you want, but for ease of debugging, you might implement the cost function and derivative computation (backpropagation) only for the squared error term first (this corresponds to setting λ = β = 0), and implement the gradient checking method in the next section to first verify that this code is correct. Then only after you have verified that the objective and derivative calculations corresponding to the squared error term are working, add in code to compute the weight decay and sparsity penalty terms and their corresponding derivatives.
Step 3: Gradient checking
Following Section 2.3 of the lecture notes, implement code for gradient checking. Specifically, complete the code incomputeNumericalGradient.m. Please use EPSILON = 10-4 as described in the lecture notes.
We've also provided code in checkNumericalGradient.m for you to test your code. This code defines a simple quadratic function 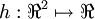 given by
given by 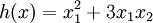 , and evaluates it at the point x = (4,10)T. It allows you to verify that your numerically evaluated gradient is very close to the true (analytically computed) gradient.
, and evaluates it at the point x = (4,10)T. It allows you to verify that your numerically evaluated gradient is very close to the true (analytically computed) gradient.
After using checkNumericalGradient.m to make sure your implementation is correct, next use computeNumericalGradient.m to make sure that yoursparseAutoencoderCost.m is computing derivatives correctly. For details, see Steps 3 in train.m. We strongly encourage you not to proceed to the next step until you've verified that your derivative computations are correct.
Implementational tip: If you are debugging your code, performing gradient checking on smaller models and smaller training sets (e.g., using only 10 training examples and 1-2 hidden units) may speed things up.
Step 4: Train the sparse autoencoder
Now that you have code that computes Jsparse and its derivatives, we're ready to minimize Jsparse with respect to its parameters, and thereby train our sparse autoencoder.
We will use the L-BFGS algorithm. This is provided to you in a function called minFunc (code provided by Mark Schmidt) included in the starter code. (For the purpose of this assignment, you only need to call minFunc with the default parameters. You do not need to know how L-BFGS works.) We have already provided code in train.m (Step 4) to call minFunc. The minFunc code assumes that the parameters to be optimized are a long parameter vector; so we will use the "θ" parameterization rather than the "(W(1),W(2),b(1),b(2))" parameterization when passing our parameters to it.
Train a sparse autoencoder with 64 input units, 25 hidden units, and 64 output units. In our starter code, we have provided a function for initializing the parameters. We initialize the biases 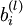 to zero, and the weights
to zero, and the weights 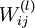 to random numbers drawn uniformly from the interval
to random numbers drawn uniformly from the interval ![\left[-\sqrt{\frac{6}{n_{\rm in}+n_{\rm out}+1}},\sqrt{\frac{6}{n_{\rm in}+n_{\rm out}+1}}\,\right]](http://deeplearning.stanford.edu/wiki/images/math/b/1/e/b1e650d9fdd8c53c515c23d49fc8fe40.png) , where nin is the fan-in (the number of inputs feeding into a node) and nout is the fan-in (the number of units that a node feeds into).
, where nin is the fan-in (the number of inputs feeding into a node) and nout is the fan-in (the number of units that a node feeds into).
The values we provided for the various parameters (λ,β,ρ, etc.) should work, but feel free to play with different settings of the parameters as well.
Implementational tip: Once you have your backpropagation implementation correctly computing the derivatives (as verified using gradient checking in Step 3), when you are now using it with L-BFGS to optimize Jsparse(W,b), make sure you're not doing gradient-checking on every step. Backpropagation can be used to compute the derivatives of Jsparse(W,b) fairly efficiently, and if you were additionally computing the gradient numerically on every step, this would slow down your program significantly.
Step 5: Visualization
After training the autoencoder, use display_network.m to visualize the learned weights. (See train.m, Step 5.) Run "print -djpeg weights.jpg" to save the visualization to a file "weights.jpg" (which you will submit together with your code).
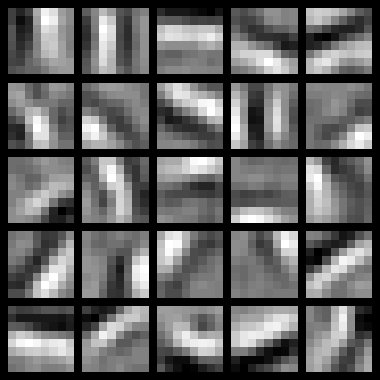
Results
To successfully complete this assignment, you should demonstrate your sparse autoencoder algorithm learning a set of edge detectors. For example, this was the visualization we obtained:
Our implementation took around 5 minutes to run on a fast computer. In case you end up needing to try out multiple implementations or different parameter values, be sure to budget enough time for debugging and to run the experiments you'll need.
Also, by way of comparison, here are some visualizations from implementations that we do not consider successful (either a buggy implementation, or where the parameters were poorly tuned):



























 3819
3819











 被折叠的 条评论
为什么被折叠?
被折叠的 条评论
为什么被折叠?








