OpenCV3.2自带的SVM不支持多线程,http://pisvm.sourceforge.net/installation.html 这个piSVM好像支持。
一、安装MPI
按照这个教程,先检查是否安装了MPI,我的电脑里没有按照,于是先下载MPI : http://www.mpich.org/downloads/ 然后按照http://www.linuxidc.com/Linux/2011-12/50236.htm 进行安装到第4步即可。接下来按照http://pisvm.sourceforge.net/installation.html 但我按照完毕 没有bin这个文件夹,于是我环境变量那里就写到mpich-3.2 再重启检查是否安装成功,but失败。原来我的make install还是要在root下安装才会生成bin文件夹。MPI别的版本:http://www.mpich.org/static/downloads/1.5/ 但依旧失败:no mpirun in
解决办法:
[root@localhost lib]# yum -y install openmpi openmpi-devel# module avail (在最大目录下打开终端 不要在刚刚安装openmpi的那个终端)dot module-cvs module-info modules null use.own
------------------------------------------------------------- /etc/modulefiles --------------------------------------------------------------
openmpi-x86_64
然后:
# module load mpi/openmpi-x86_64 输入 which mpirun 即可看到一个路径。成功!
MPI 相关命令的资料 http://linuxcommand.org/man_pages/mpirun1.html
https://docs.oracle.com/cd/E19708-01/821-1319-10/ExecutingPrograms.html
二、安装测试piSVM
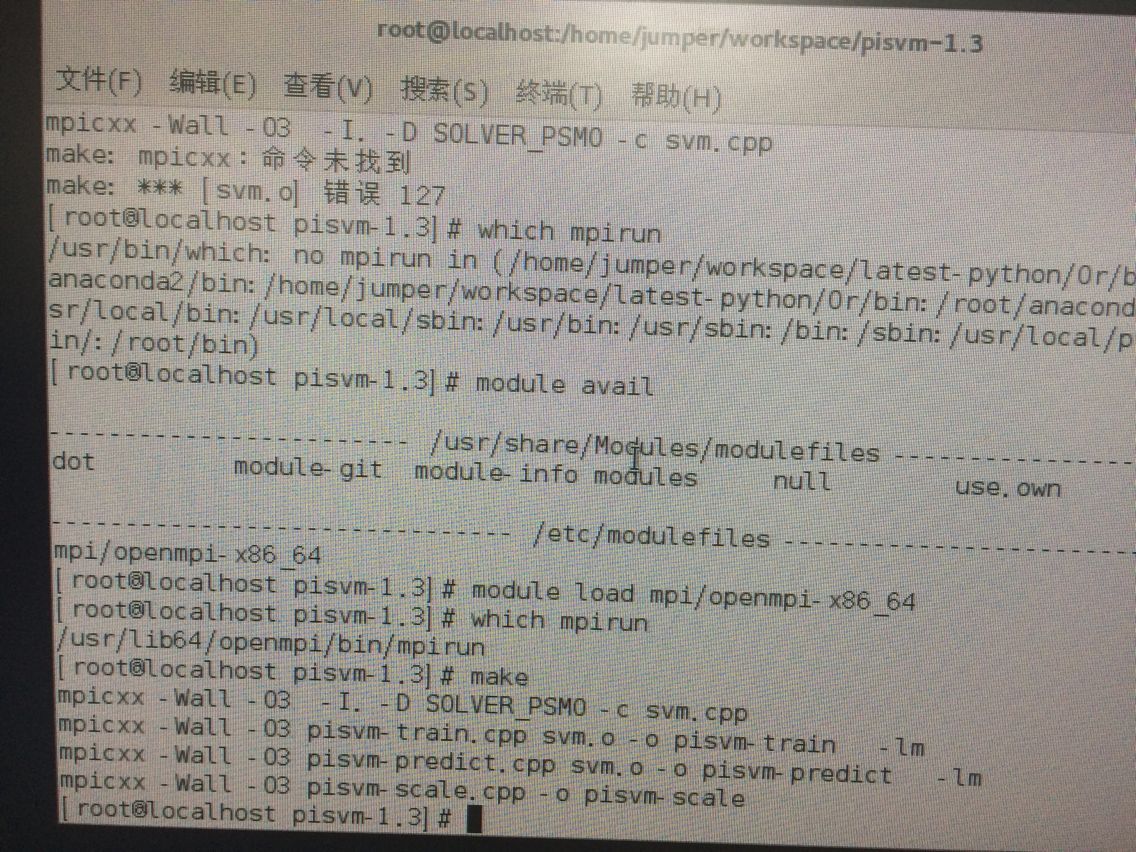
本来按照 http://pisvm.sourceforge.net/installation.html 在装完MPI 以后就下载piSVM然后make 结果又提示找不到mpi了 原来又要
module load mpi/openmpi-x86_64 如:
即可。
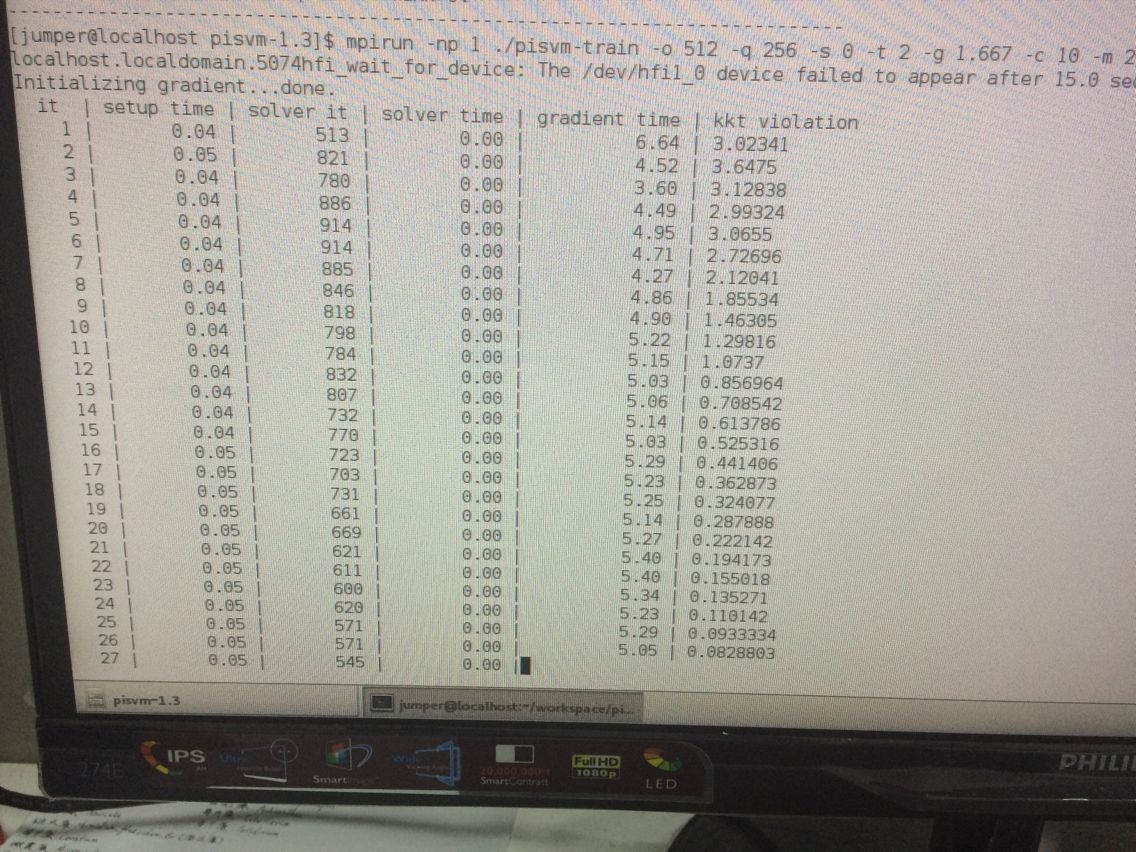
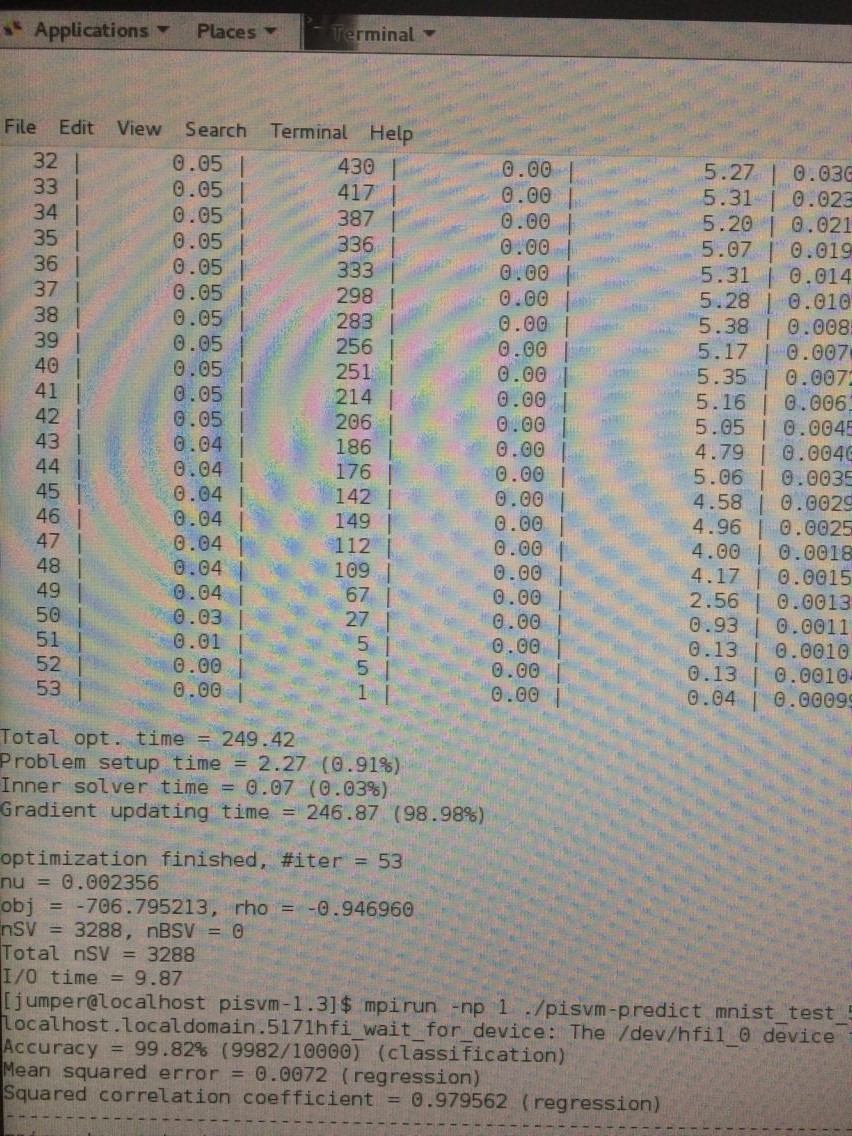
接下来开始测试piSVM 在:mpirun -np 1 ./pisvm-train -o 512 -q 256 -s 0 -t 2 -g 1.667 -c 10 -m 256 mnist_train_576_rbf_8vr.dat
出现报错:...You can override this protection by adding the --allow-run-as-root option to your cmd line... 我在这句后面加上--allow-run-as-root还是一样的报错。尝试不root启动,而是user重启电脑,即可:
在正常训练,会生成model文件。
预测分类部分:
初步成功!!!
三、多线程的SVM
1、多线程小例子:
#include <iostream>
#include <cstdlib>
#include <pthread.h>
#include <boost/thread.hpp>
using namespace std;
#define NUM_THREADS 4
void *PrintHello(void *threadID)
{
long tid;
tid=(long)threadID;
cout<<"hello world,Thread ID: "<<tid<<endl;
sleep(100);
for(int c=0;c<8000000;c++)
{
long k=c*c;
long j=c+c;
for(int cc=0;cc<8000000;cc++)
{
long kk=cc*2;
}
}
pthread_exit(NULL);
}
int main()
{
pthread_t threads[NUM_THREADS];
int rc;
int i;
for(i=0;i<NUM_THREADS;i++)
{
cout<<"main():create thread: "<<i<<endl;
rc=pthread_create(&threads[i],NULL,PrintHello,(void*)i);
if(rc)
{
cout<<"Error:Unable to create thread, "<<rc<<endl;
exit(-1);
}
}
for(i=0;i<NUM_THREADS;i++)
{
pthread(threads[i],NULL);
}
pthread_exit(NULL);
return 0;
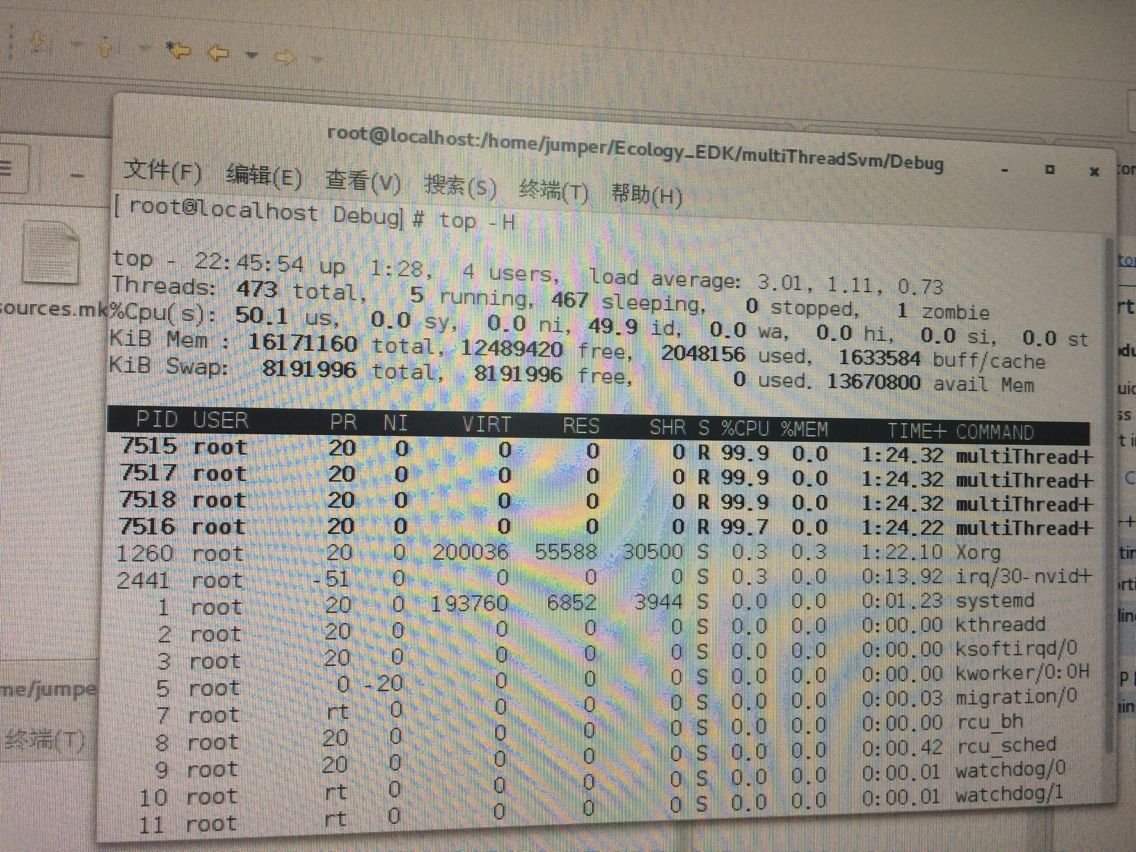
}
2、piSVM--单进程下单线程测试
这个piSVM 看了下它的predict.cpp源码 里面仍旧是基于MPI库的,用终端通过MPI的相关命令进行预测分类已经测试过了。现在不通过MPI进行预测分类,而是写成一个普通的测试分类文件:
#include <stdio.h>
#include <ctype.h>
#include <stdlib.h>
#include <string.h>
#include <errno.h>
#include "svm.h"
int print_null(const char *s,...) {return 0;}
static int (*info)(const char *fmt,...) = &printf;
struct svm_node *x;
int max_nr_attr = 64;
struct svm_model* model;
int predict_probability=0;
static char *line = NULL;
static int max_line_len;
static char* readline(FILE *input)
{
int len;
if(fgets(line,max_line_len,input) == NULL)
return NULL;
while(strrchr(line,'\n') == NULL)
{
max_line_len *= 2;
line = (char *) realloc(line,max_line_len);
len = (int) strlen(line);
if(fgets(line+len,max_line_len-len,input) == NULL)
break;
}
return line;
}
void exit_input_error(int line_num)
{
fprintf(stderr,"Wrong input format at line %d\n", line_num);
exit(1);
}
void predict(FILE *input, FILE *output)
{
int correct = 0;
int total = 0;
double error = 0;
double sump = 0, sumt = 0, sumpp = 0, sumtt = 0, sumpt = 0;
int svm_type=svm_get_svm_type(model);
int nr_class=svm_get_nr_class(model);
double *prob_estimates=NULL;
int j;
if(predict_probability)
{
if (svm_type==NU_SVR || svm_type==EPSILON_SVR)
info("Prob. model for test data: target value = predicted value + z,\nz: Laplace distribution e^(-|z|/sigma)/(2sigma),sigma=%g\n",svm_get_svr_probability(model));
else
{
int *labels=(int *) malloc(nr_class*sizeof(int));
svm_get_labels(model,labels);
prob_estimates = (double *) malloc(nr_class*sizeof(double));
fprintf(output,"labels");
for(j=0;j<nr_class;j++)
fprintf(output," %d",labels[j]);
fprintf(output,"\n");
free(labels);
}
}
max_line_len = 1024;
line = (char *)malloc(max_line_len*sizeof(char));
while(readline(input) != NULL)
{
int i = 0;
double target_label, predict_label;
char *idx, *val, *label, *endptr;
int inst_max_index = -1; // strtol gives 0 if wrong format, and precomputed kernel has <index> start from 0
label = strtok(line," \t\n");
if(label == NULL) // empty line
exit_input_error(total+1);
target_label = strtod(label,&endptr);
if(endptr == label || *endptr != '\0')
exit_input_error(total+1);
while(1)
{
if(i>=max_nr_attr-1) // need one more for index = -1
{
max_nr_attr *= 2;
x = (struct svm_node *) realloc(x,max_nr_attr*sizeof(struct svm_node));
}
idx = strtok(NULL,":");
val = strtok(NULL," \t");
if(val == NULL)
break;
errno = 0;
x[i].index = (int) strtol(idx,&endptr,10);
if(endptr == idx || errno != 0 || *endptr != '\0' || x[i].index <= inst_max_index)
exit_input_error(total+1);
else
inst_max_index = x[i].index;
errno = 0;
x[i].value = strtod(val,&endptr);
if(endptr == val || errno != 0 || (*endptr != '\0' && !isspace(*endptr)))
exit_input_error(total+1);
++i;
}
x[i].index = -1;
if (predict_probability && (svm_type==C_SVC || svm_type==NU_SVC))
{
predict_label = svm_predict_probability(model,x,prob_estimates);
fprintf(output,"%g",predict_label);
for(j=0;j<nr_class;j++)
fprintf(output," %g",prob_estimates[j]);
fprintf(output,"\n");
}
else
{
predict_label = svm_predict(model,x);
fprintf(output,"%g\n",predict_label);
}
if(predict_label == target_label)
++correct;
error += (predict_label-target_label)*(predict_label-target_label);
sump += predict_label;
sumt += target_label;
sumpp += predict_label*predict_label;
sumtt += target_label*target_label;
sumpt += predict_label*target_label;
++total;
}
if (svm_type==NU_SVR || svm_type==EPSILON_SVR)
{
info("Mean squared error = %g (regression)\n",error/total);
info("Squared correlation coefficient = %g (regression)\n",
((total*sumpt-sump*sumt)*(total*sumpt-sump*sumt))/
((total*sumpp-sump*sump)*(total*sumtt-sumt*sumt))
);
}
else
info("Accuracy = %g%% (%d/%d) (classification)\n",
(double)correct/total*100,correct,total);
info("Mean squared error = %g (regression)\n",error/total);
info("Squared correlation coefficient = %g (regression)\n",
((total*sumpt-sump*sumt)*(total*sumpt-sump*sumt))/
((total*sumpp-sump*sump)*(total*sumtt-sumt*sumt))
);
if(predict_probability)
free(prob_estimates);
}
void exit_with_help()
{
printf(
"Usage: svm-predict [options] test_file model_file output_file\n"
"options:\n"
"-b probability_estimates: whether to predict probability estimates, 0 or 1 (default 0); for one-class SVM only 0 is supported\n"
"-q : quiet mode (no outputs)\n"
);
exit(1);
}
int main(int argc, char **argv)
{
FILE *input, *output;
input = fopen("mnist_test_576_rbf_8vr.dat","r");
if(input == NULL)
{
fprintf(stderr,"can't open input file \n");
exit(1);
}
output = fopen("out0629_1.txt","w");
if(output == NULL)
{
fprintf(stderr,"can't open output file\n");
exit(1);
}
if((model=svm_load_model("mnist_train_576_rbf_8vr.dat.model"))==0)
{
fprintf(stderr,"can't open model file \n");
exit(1);
}
x = (struct svm_node *) malloc(max_nr_attr*sizeof(struct svm_node));
if(predict_probability)
{
if(svm_check_probability_model(model)==0)
{
fprintf(stderr,"Model does not support probabiliy estimates\n");
exit(1);
}
}
else
{
if(svm_check_probability_model(model)!=0)
info("Model supports probability estimates, but disabled in prediction.\n");
}
predict(input,output);
svm_free_and_destroy_model(&model);
free(x);
free(line);
fclose(input);
fclose(output);
return 0;
}3、多线程下的piSVM
#include <stdio.h>
#include <ctype.h>
#include <stdlib.h>
#include <string.h>
#include <errno.h>
#include "svm.h"
#include <iostream>
#include <cstdlib>
#include <pthread.h>
#include <boost/thread.hpp>
using namespace std;
#define NUM_THREADS 4
int print_null(const char *s,...) {return 0;}
static int (*info)(const char *fmt,...) = &printf;
struct svm_node *x;
struct svm_model* model;
struct threadsArgs
{
FILE *inputfile;
FILE *outputfile;
svm_model *modelfile;
};
struct threadsArgs arg;
int max_nr_attr = 64;
int predict_probability=0;
static char *line = NULL;
static int max_line_len;
static char* readline(FILE *input)
{
int len;
if(fgets(line,max_line_len,input) == NULL)
return NULL;
while(strrchr(line,'\n') == NULL)
{
max_line_len *= 2;
line = (char *) realloc(line,max_line_len);
len = (int) strlen(line);
if(fgets(line+len,max_line_len-len,input) == NULL)
break;
}
return line;
}
void exit_input_error(int line_num)
{
fprintf(stderr,"Wrong input format at line %d\n", line_num);
exit(1);
}
void predict(FILE *input, FILE *output,svm_model *model)
{
cout<<"begin to enter predict()"<<endl;
int correct = 0;
int total = 0;
double error = 0;
double sump = 0, sumt = 0, sumpp = 0, sumtt = 0, sumpt = 0;
int svm_type=svm_get_svm_type(model);
int nr_class=svm_get_nr_class(model);
double *prob_estimates=NULL;
int j;
if(predict_probability)
{
if (svm_type==NU_SVR || svm_type==EPSILON_SVR)
info("Prob. model for test data: target value = predicted value + z,\nz: Laplace distribution e^(-|z|/sigma)/(2sigma),sigma=%g\n",svm_get_svr_probability(model));
else
{
int *labels=(int *) malloc(nr_class*sizeof(int));
svm_get_labels(model,labels);
prob_estimates = (double *) malloc(nr_class*sizeof(double));
fprintf(output,"labels");
for(j=0;j<nr_class;j++)
fprintf(output," %d",labels[j]);
fprintf(output,"\n");
free(labels);
}
}
max_line_len = 1024;
line = (char *)malloc(max_line_len*sizeof(char));
while(readline(input) != NULL)
{
int i = 0;
double target_label, predict_label;
char *idx, *val, *label, *endptr;
int inst_max_index = -1; // strtol gives 0 if wrong format, and precomputed kernel has <index> start from 0
label = strtok(line," \t\n");
if(label == NULL) // empty line
exit_input_error(total+1);
target_label = strtod(label,&endptr);
if(endptr == label || *endptr != '\0')
exit_input_error(total+1);
while(1)
{
if(i>=max_nr_attr-1) // need one more for index = -1
{
max_nr_attr *= 2;
x = (struct svm_node *) realloc(x,max_nr_attr*sizeof(struct svm_node));
}
idx = strtok(NULL,":");
val = strtok(NULL," \t");
if(val == NULL)
break;
errno = 0;
x[i].index = (int) strtol(idx,&endptr,10);
if(endptr == idx || errno != 0 || *endptr != '\0' || x[i].index <= inst_max_index)
exit_input_error(total+1);
else
inst_max_index = x[i].index;
errno = 0;
x[i].value = strtod(val,&endptr);
if(endptr == val || errno != 0 || (*endptr != '\0' && !isspace(*endptr)))
exit_input_error(total+1);
++i;
}
x[i].index = -1;
if (predict_probability && (svm_type==C_SVC || svm_type==NU_SVC))
{
predict_label = svm_predict_probability(model,x,prob_estimates);
fprintf(output,"%g",predict_label);
for(j=0;j<nr_class;j++)
fprintf(output," %g",prob_estimates[j]);
fprintf(output,"\n");
}
else
{
predict_label = svm_predict(model,x);
fprintf(output,"%g\n",predict_label);
}
if(predict_label == target_label)
++correct;
error += (predict_label-target_label)*(predict_label-target_label);
sump += predict_label;
sumt += target_label;
sumpp += predict_label*predict_label;
sumtt += target_label*target_label;
sumpt += predict_label*target_label;
++total;
}
if (svm_type==NU_SVR || svm_type==EPSILON_SVR)
{
info("Mean squared error = %g (regression)\n",error/total);
info("Squared correlation coefficient = %g (regression)\n",
((total*sumpt-sump*sumt)*(total*sumpt-sump*sumt))/
((total*sumpp-sump*sump)*(total*sumtt-sumt*sumt))
);
}
else
{
info("Accuracy = %g%% (%d/%d) (classification)\n",
(double)correct/total*100,correct,total);
info("Mean squared error = %g (regression)\n",error/total);
info("Squared correlation coefficient = %g (regression)\n",
((total*sumpt-sump*sumt)*(total*sumpt-sump*sumt))/
((total*sumpp-sump*sump)*(total*sumtt-sumt*sumt))
);
}
if(predict_probability)
free(prob_estimates);
}
void exit_with_help()
{
printf(
"Usage: svm-predict [options] test_file model_file output_file\n"
"options:\n"
"-b probability_estimates: whether to predict probability estimates, 0 or 1 (default 0); for one-class SVM only 0 is supported\n"
"-q : quiet mode (no outputs)\n"
);
exit(1);
}
void *mainPredict(void *arg)
{
//struct threadsArgs *pstru;
//pstru=(struct threadsArgs*)arg;
threadsArgs *pstru;
pstru=(struct threadsArgs *)arg;
x = (struct svm_node *) malloc(max_nr_attr*sizeof(struct svm_node));
if(predict_probability)
{
if(svm_check_probability_model(pstru->modelfile)==0)
{
fprintf(stderr,"Model does not support probabiliy estimates\n");
exit(1);
}
}
else
{
if(svm_check_probability_model(pstru->modelfile)!=0)
info("Model supports probability estimates, but disabled in prediction.\n");
}
//FILE *theInput=pstru->inputfile;
predict(pstru->inputfile,pstru->outputfile,pstru->modelfile);
free(x);
free(line);
//svm_free_and_destroy_model(&pstru->modelfile);
//fclose(pstru->inputfile);
//fclose(pstru->outputfile);
}
int main()
{
pthread_t threads[NUM_THREADS];
int rc;
//the first thread:
int i=0;
cout<<"main():create thread: "<<i<<endl;
struct threadsArgs arg0;
arg0.inputfile=fopen("/home/jumper/Ecology_EDK/multiThreadSvm/mnist_test_576_rbf_8vr.dat","r");
arg0.outputfile=fopen("/home/jumper/Ecology_EDK/multiThreadSvm/out0629_1.txt","w");
arg0.modelfile=svm_load_model("/home/jumper/Ecology_EDK/multiThreadSvm/mnist_train_576_rbf_8vr.dat.model");
rc=pthread_create(&threads[i],NULL,mainPredict,&arg0);
if(rc)
{
cout<<"Error:Unable to create thread, "<<rc<<endl;
exit(-1);
}
//the second thread:
i=1;
cout<<"main():create thread: "<<i<<endl;
struct threadsArgs arg1;
arg1.inputfile=fopen("/home/jumper/Ecology_EDK/multiThreadSvm/mnist_test_576_rbf_8vr.dat","r");
arg1.outputfile=fopen("/home/jumper/Ecology_EDK/multiThreadSvm/out0629_2.txt","w");
//arg1.modelfile=svm_load_model("mnist_train_576_rbf_8vr.dat.model");
arg1.modelfile=arg0.modelfile;
rc=pthread_create(&threads[i],NULL,mainPredict,&arg1);
if(rc)
{
cout<<"Error:Unable to create thread, "<<rc<<endl;
exit(-1);
}
//the third thread:
i=2;
cout<<"main():create thread: "<<i<<endl;
struct threadsArgs arg2;
arg2.inputfile=fopen("/home/jumper/Ecology_EDK/multiThreadSvm/mnist_test_576_rbf_8vr.dat","r");
arg2.outputfile=fopen("/home/jumper/Ecology_EDK/multiThreadSvm/out0629_3.txt","w");
//arg2.modelfile=svm_load_model("mnist_train_576_rbf_8vr.dat.model");
arg2.modelfile=arg0.modelfile;
rc=pthread_create(&threads[i],NULL,mainPredict,&arg2);
if(rc)
{
cout<<"Error:Unable to create thread, "<<rc<<endl;
exit(-1);
}
//the third thread:
i=3;
cout<<"main():create thread: "<<i<<endl;
struct threadsArgs arg3;
arg3.inputfile=fopen("/home/jumper/Ecology_EDK/multiThreadSvm/mnist_test_576_rbf_8vr.dat","r");
arg3.outputfile=fopen("/home/jumper/Ecology_EDK/multiThreadSvm/out0629_4.txt","w");
//arg3.modelfile=svm_load_model("mnist_train_576_rbf_8vr.dat.model");
arg3.modelfile=arg0.modelfile;
rc=pthread_create(&threads[i],NULL,mainPredict,&arg3);
if(rc)
{
cout<<"Error:Unable to create thread, "<<rc<<endl;
exit(-1);
}
pthread_join(threads[0],NULL);
pthread_join(threads[1],NULL);
pthread_join(threads[2],NULL);
pthread_join(threads[3],NULL);
//pthread_exit(NULL);
//svm_free_and_destroy_model(&arg.modelfile);
//fclose(arg.inputfile);
//fclose(arg.outputfile);
return 0;
}
看了下网上查到的另几个资料也和这个这不多,如果这个没戏 另几个应该也没戏:
https://github.com/ewalker544/libsvm_task
https://github.com/nagyistoce/opencl-libsvm
https://github.com/ewalker544/libsvm_threads
https://github.com/markodjurovic/parallelLibSvm
https://github.com/henryzord/libSVM-OpenCL
https://code.google.com/archive/p/psvm/downloads
到底哪里出了问题 多线程的piSVM。。。好烦躁
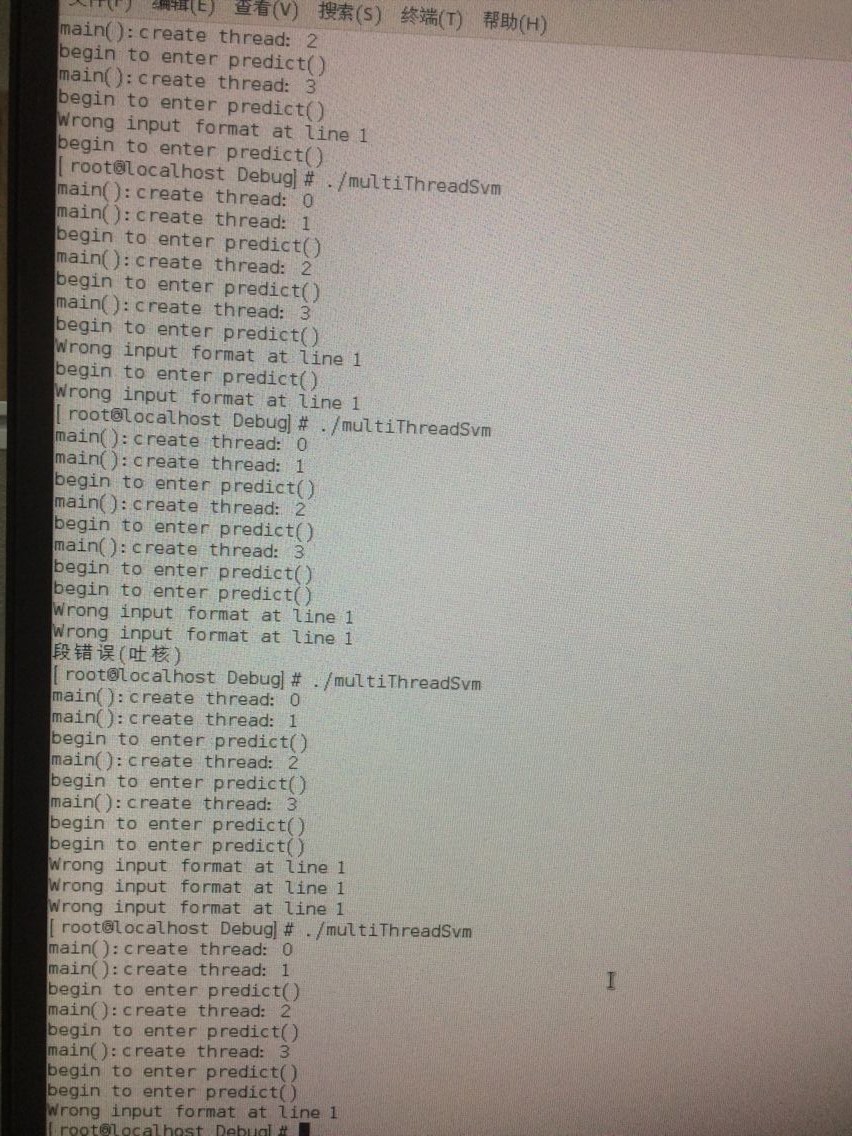
哦原来是使用了一个在多线程下不安全的函数 :strtok() !!!这也就是说即使改成不依赖MPI的cpp,依旧是不支持多线程的,因为它这个SVM作者在写时里面用的都是单线程安全的函数,根本考虑的不是多线程下的SVM!!!所以这个piSVM其实是只支持多进程 不支持多线程!!!!!!!!
还是想再改一下,将线程不安全的函数改成支持多线程的函数,看是否可以:
将刚刚的strtok() 按照http://blog.csdn.net/hustfoxy/article/details/23473805 改成strtok_r() !但依旧吐核?!
四、XGBoost
同事说 xgboost支持多线程 而且很强大 比SVM快3倍,而且准确率不会差!https://github.com/dmlc/xgboost 已经在windows和linux下有人测试过了,的确如此!
https://blog.csdn.net/peterchan88/article/details/77921803 原理
https://mp.weixin.qq.com/s?__biz=MzUyMjE2MTE0Mw==&mid=2247484525&idx=1&sn=e554d0ac010092c9d2c24ae57d279c59&chksm=f9d15af5cea6d3e3fcb8056aeea7d682f3e496e3b7394cbbefcad6ad8c7fb43fcd2d69e056cf&scene=21#wechat_redirect 参数意义
http://www.360doc.com/content/17/1123/17/35874779_706497143.shtml 参数意义
https://blog.csdn.net/szu_hadooper/article/details/79050483 模型保存读取
https://blog.csdn.net/zc02051126/article/details/46709599 二分类
https://blog.csdn.net/zc02051126/article/details/46771793
https://www.2cto.com/kf/201607/528771.html 参数调优
https://www.cnblogs.com/harekizgel/p/7683803.html 输出参数的意义
https://blog.csdn.net/u011089523/article/details/72812019 手写数字 多分类
http://www.360doc.com/content/17/1123/17/35874779_706497143.shtml 分类和回归
http://xgboost.readthedocs.io/en/latest/python/python_intro.html 官网介绍
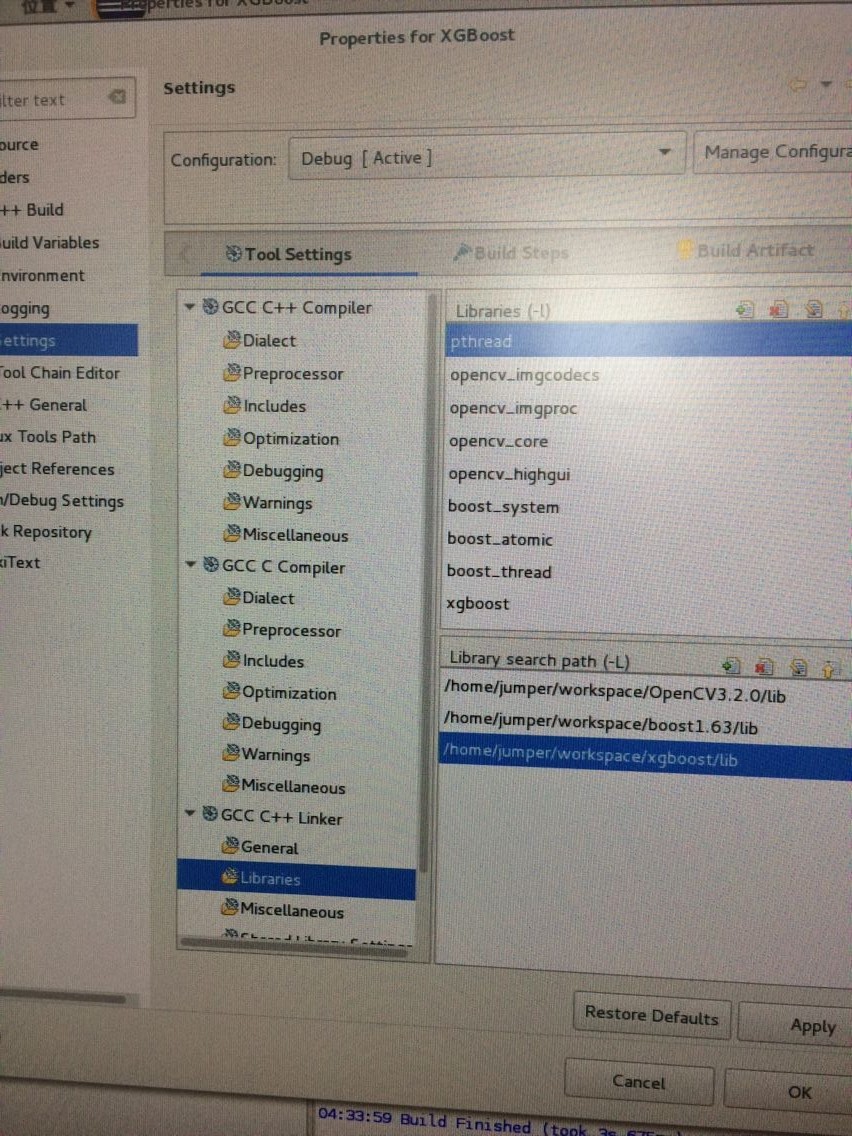
C++使用方式:
编译好后,
#ifndef SRC_GHUTEGETIMAGE_THREAD_H_
#define SRC_GHUTEGETIMAGE_THREAD_H_
#include <opencv2/opencv.hpp>
#include <opencv2/imgproc/imgproc.hpp>
#include <opencv2/highgui/highgui.hpp>
#include <iostream>
#include <sys/types.h>
#include <dirent.h>
#include <sys/stat.h>
#include <iterator>
#include <unistd.h>
#include <c_api.h>
using namespace cv;
using namespace std;
class GhuteGetImage_Thread{
public:
GhuteGetImage_Thread(unsigned char);
void threadrun();
int WriteData(string, cv::Mat&);
int LoadData(string, cv::Mat&, int, int, int);
~GhuteGetImage_Thread();
private:
int m_chuteorder;
bool m_threadrun;
Mat m_features;
BoosterHandle h_booster;
};
#endif /* SRC_GHUTEGETIMAGE_THREAD_H_ */#include "GhuteGetImage_Thread.h"
GhuteGetImage_Thread::GhuteGetImage_Thread(unsigned char _chuteor)
:m_threadrun(true)
{
// TODO Auto-generated constructor stub
m_chuteorder=_chuteor;
int ret=XGBoosterCreate(0,0,&h_booster);
printf("Create ret=%d\n",ret);
ret=XGBoosterLoadModel(h_booster,"/home/jumper/Ecology_EDK/XgboostTest/YinZao/xgb_yinzao.model");
printf("LoadModel ret=%d\n",ret);
ret=LoadData("/home/jumper/Ecology_EDK/XgboostTest/YinZao/feature.txt",m_features,19735,1062, 1);
printf("ret=%d rows=%d cold=%d\n",ret,m_features.rows,m_features.cols);
}
GhuteGetImage_Thread::~GhuteGetImage_Thread() {
// TODO Auto-generated destructor stub
XGBoosterFree(h_booster);
}
/*----------------------------
* 功能 : 从 .txt 文件中读入数据,保存到 cv::Mat 矩阵
* - 默认按 float 格式读入数据,
* - 如果没有指定矩阵的行、列和通道数,则输出的矩阵是单通道、N 行 1 列的
*----------------------------
* 函数 : LoadData
* 访问 : public
* 返回 : -1:打开文件失败;0:按设定的矩阵参数读取数据成功;1:按默认的矩阵参数读取数据
*
* 参数 : fileName [in] 文件名
* 参数 : matData [out] 矩阵数据
* 参数 : matRows [in] 矩阵行数,默认为 0
* 参数 : matCols [in] 矩阵列数,默认为 0
* 参数 : matChns [in] 矩阵通道数,默认为 0
*/
int GhuteGetImage_Thread::LoadData(string fileName, cv::Mat& matData, int matRows = 0, int matCols = 0, int matChns = 0)
{
int retVal = 0;
// 打开文件
ifstream inFile(fileName.c_str(), ios_base::in);
if(!inFile.is_open())
{
cout << "读取文件失败" << endl;
retVal = -1;
return (retVal);
}
// 载入数据
istream_iterator<float> begin(inFile); //按 float 格式取文件数据流的起始指针
istream_iterator<float> end; //取文件流的终止位置
vector<float> inData(begin,end); //将文件数据保存至 std::vector 中
cv::Mat tmpMat = cv::Mat(inData); //将数据由 std::vector 转换为 cv::Mat
// 输出到命令行窗口
//copy(vec.begin(),vec.end(),ostream_iterator<double>(cout,"\t"));
// 检查设定的矩阵尺寸和通道数
size_t dataLength = inData.size();
//1.通道数
if (matChns == 0)
{
matChns = 1;
}
//2.行列数
if (matRows != 0 && matCols == 0)
{
matCols = dataLength / matChns / matRows;
}
else if (matCols != 0 && matRows == 0)
{
matRows = dataLength / matChns / matCols;
}
else if (matCols == 0 && matRows == 0)
{
matRows = dataLength / matChns;
matCols = 1;
}
//3.数据总长度
if (dataLength != (matRows * matCols * matChns))
{
cout << "读入的数据长度 不满足 设定的矩阵尺寸与通道数要求,将按默认方式输出矩阵!" << endl;
retVal = 1;
matChns = 1;
matRows = dataLength;
}
// 将文件数据保存至输出矩阵
matData = tmpMat.reshape(matChns, matRows).clone();
return (retVal);
}
/*----------------------------
* 功能 : 将 cv::Mat 数据写入到 .txt 文件
*----------------------------
* 函数 : WriteData
* 访问 : public
* 返回 : -1:打开文件失败;0:写入数据成功;1:矩阵为空
*
* 参数 : fileName [in] 文件名
* 参数 : matData [in] 矩阵数据
*/
int GhuteGetImage_Thread::WriteData(string fileName, cv::Mat& matData)
{
int retVal = 0;
// 检查矩阵是否为空
if (matData.empty())
{
cout << "矩阵为空" << endl;
retVal = 1;
return (retVal);
}
// 打开文件
ofstream outFile(fileName.c_str(), ios_base::out); //按新建或覆盖方式写入
if (!outFile.is_open())
{
cout << "打开文件失败" << endl;
retVal = -1;
return (retVal);
}
// 写入数据
for (int r = 0; r < matData.rows; r++)
{
for (int c = 0; c < matData.cols; c++)
{
int data = matData.at<uchar>(r,c); //读取数据,at<type> - type 是矩阵元素的具体数据格式
outFile << data << "\t" ; //每列数据用 tab 隔开
}
outFile << endl; //换行
}
return (retVal);
}
void GhuteGetImage_Thread::threadrun(){
char contxet[10]={0};
float f_testdata[1062]={0.0f};
sprintf(contxet,"result%d",m_chuteorder);
FILE* m_fp;
m_fp=fopen(contxet, "w+");
if( m_fp == NULL ){
fclose(m_fp);
m_fp=NULL;
}
printf("thread%u start\n",m_chuteorder);
int index(0);
while(m_threadrun)
{
usleep(200);
if(index<19735)
{
for(int i=0;i<1062;i++)
{
f_testdata[i]=m_features.at<float>(index,i);
}
bst_ulong out_len;
const float *f;
DMatrixHandle h_test;
int ret=XGDMatrixCreateFromMat((const float *)f_testdata, 1,1062, -1, &h_test);
ret=XGBoosterPredict(h_booster,h_test,0,0,&out_len,&f);
if(m_fp != NULL)
{
float f_data=f[0];
sprintf(contxet,"%f\n",f_data);
fwrite(contxet,sizeof(char),strlen(contxet),m_fp);
}
index++;
}
else
{
if(m_fp != NULL)
{
fclose(m_fp);
m_fp=NULL;
}
printf("thread%d finish!\n",m_chuteorder);
break;
}
}
//int nouse=XGDMatrixFree((void*)f_testdata);
}
#include "GhuteGetImage_Thread.h"
int main()
{
int threads=4;
GhuteGetImage_Thread xgboostTester(threads);
xgboostTester.threadrun();
return 0;
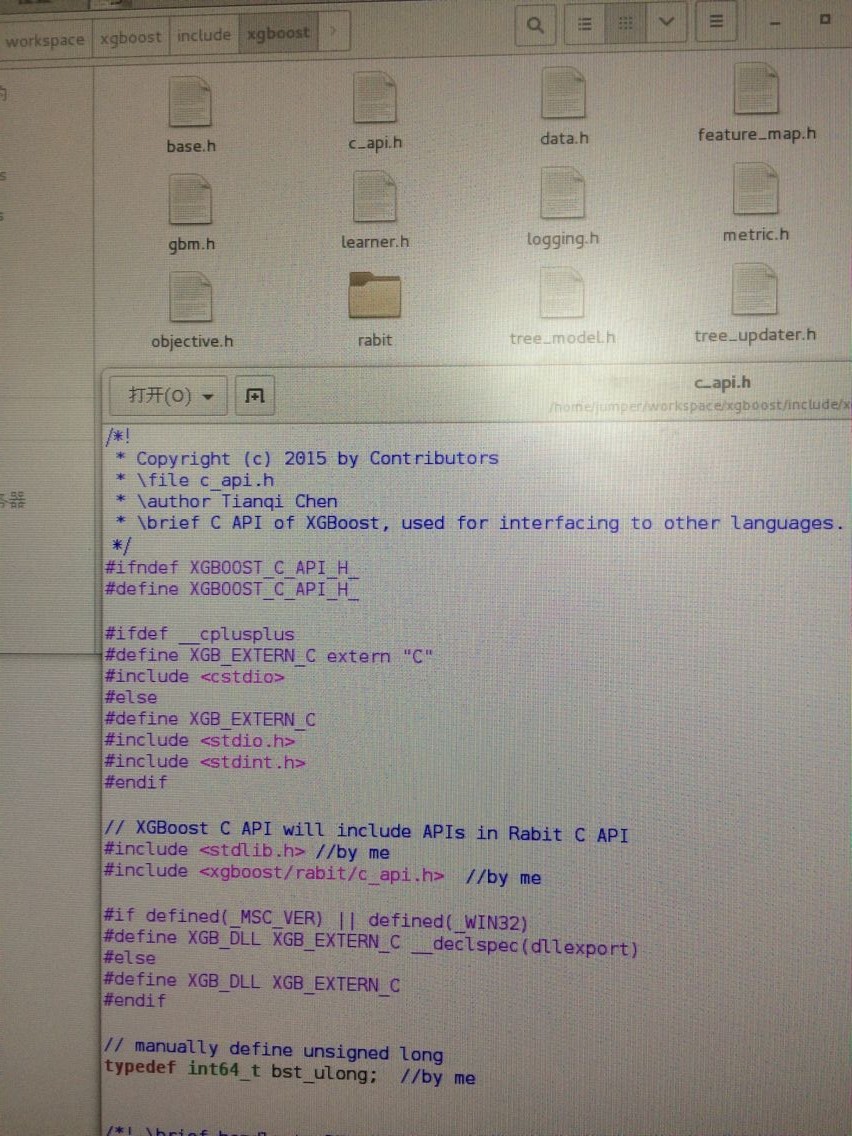
}于是我想多线程试一下:工程在:http://download.csdn.net/detail/wd1603926823/9926482 但是编译运行之前要先改一个地方(因为会出现类似: bst_ulong does not name a type! uint64_t does not name a type!之类的错误) 解决办法是:先将rabit文件夹复制到这里,然后打开这个c_api.h 修改3处 by me的地方即可。
配置部分:
结果: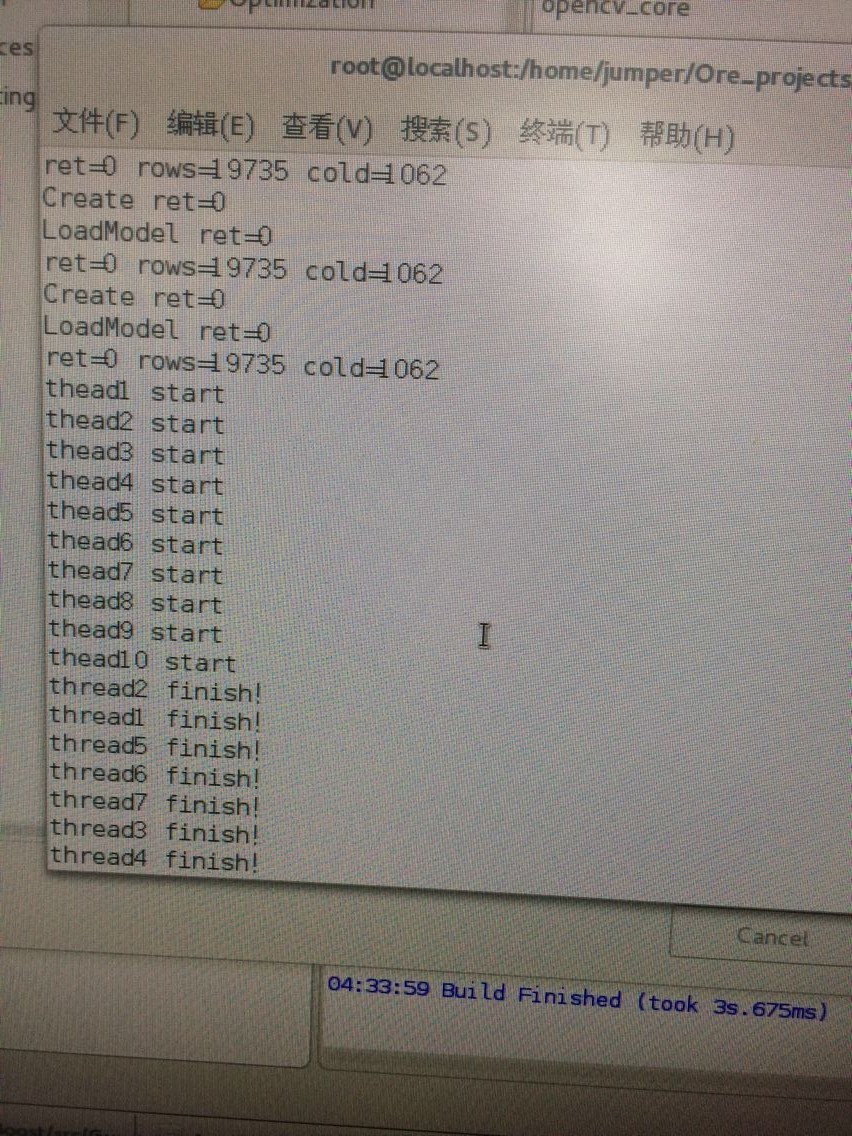
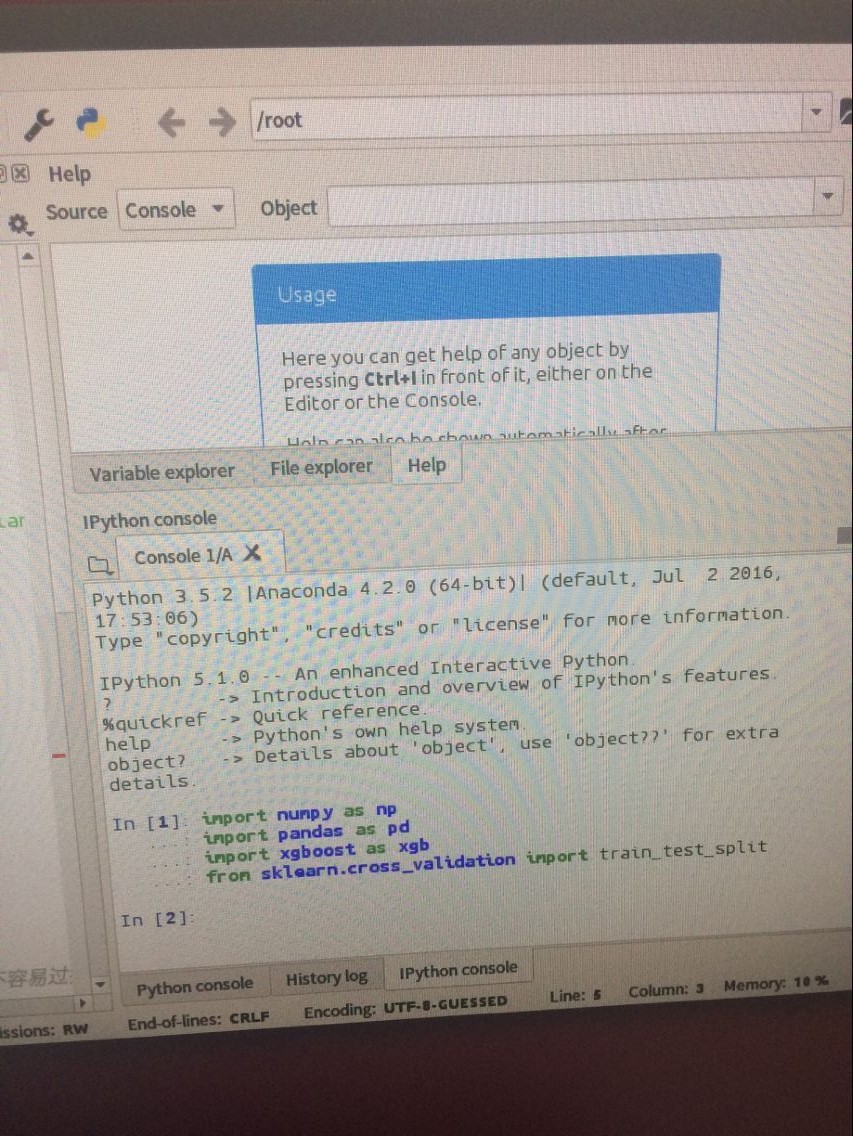
python使用方式:
今天是2018.5.10日,要在ubuntu16.04下使用XGBoost的Python接口。打开XGBoost安装目录,找到python_package,打开终端: python setup.py install 或者python3 setup.py install
然后我import xgboost时出现 错误:
1、libgomp.so.1: version `GOMP_4.0' not found (required by
解决办法:在系统内找libgomp.so.1,可能出现几个地方,即几个不同的path下都有这个名字的库,然后对每个path(根据自己的路径来)
strings path/libgomp.so.1 | grep GOMP_4.0 如果有哪个path(比如path2)的libgomp.so.1出现了GOMP_4.0
就将其它路径下的libgomp.so.1删掉 然后将path2下的libgomp.so.1 复制即通过终端cp到另几个path下即可
2、libstdc++.so.6: version `GLIBCXX_3.4.20' not found
解决办法:与上面那个错误的解决办法类似。依旧是去找系统中libstdc++.so.6 (我是CTRL+F查找的),然后可以看到不同路径path下
的libstdc++.so.6,依旧是去查看来自不同path的libstdc++.so.6有哪个有GLIBCXX_3.4.20,
即strings path/libstdc++.so.6 | grep GLIBCXX_3.4.20 如果找到是path3下满足要求,就将别的path下的libstdc++.so.6删掉,
然后将path3下的libstdc++.so.6 复制 cp过去即可 现在已经没问题了。可以使用了不会再报错。
对于XGBoost的python接口的使用,先自己生成自己想要的特征:
//生成特征供xgboost使用
int main()
{
//...此处代码不能显示
FILE *txtfile = fopen("/home/jumper/Ecology_projects/Algaes_train_test/zhilianzao/train.csv", "w");
bool istrain=true;
//set column name for every column...
if(istrain==true)
{
fprintf(txtfile, "Label,");
}
for (int col_ind_pos = 0; col_ind_pos != DIM_OF_SAMPLE96X32-1; col_ind_pos++)
{
fprintf(txtfile, "%d,",col_ind_pos);
}
fprintf(txtfile, "%d\n",DIM_OF_SAMPLE96X32-1);
if(istrain==true)
{
//训练样本
for (int img_ind_pos = 0; img_ind_pos != 384; img_ind_pos++)
{
sprintf(srcimgs, "/home/jumper/Ecology_projects/Algaes_train_test/zhilianzao/test/positive/%d.JPG", img_ind_pos);
Mat src = imread(srcimgs,0);
if(!src.data)
{
continue;
}
cout<<"extract feature of image---"<<img_ind_pos<<"..."<<endl;
//...此处代码不能显示
//生成训练样本的正样本的特征和标签
if(istrain==true)
{
int label = 1;
fprintf(txtfile, "%d,", label); //use "," instead of "\t"
}
float f_testdata[DIM_OF_SAMPLE96X32] = { 0.0f };
for (int i = 0; i < DIM_OF_SAMPLE96X32; i++)
{
if (i < DIM_OF_HOG96X32)
{
f_testdata[i] = descriptors0[i];
fprintf(txtfile, "%f,", descriptors0[i]);
}
else if (i<DIM_OF_HOG96X32+DIM_OF_LBP1)
{
f_testdata[i] = LBPH00.at<float>(0, i - DIM_OF_HOG96X32);
fprintf(txtfile, "%f,", LBPH00.at<float>(0, i - DIM_OF_HOG96X32));
}
else if (i == DIM_OF_SAMPLE96X32-2)
{
f_testdata[i] = (float)src.rows*src.cols / 1000.0f;
fprintf(txtfile, "%f,", (float)src.rows*src.cols / 1000.0f);
}
else
{
f_testdata[i] = max((float)src.rows / (float)src.cols, (float)src.cols / (float)src.rows);
fprintf(txtfile, "%f\n", max((float)src.rows / (float)src.cols, (float)src.cols / (float)src.rows));
}
}
//train positive features...
}
for (int img_ind_pos = 0; img_ind_pos != 210; img_ind_pos++)
{
sprintf(srcimgs, "/home/jumper/Ecology_projects/Algaes_train_test/zhilianzao/test/negative/%d.JPG", img_ind_pos);
Mat src = imread(srcimgs,0);
if(!src.data)
{
continue;
}
cout<<"extract feature of image---"<<img_ind_pos<<"..."<<endl;
//...此处代码不能显示
//生成训练样本的负样本的特征和标签
if(istrain==true)
{
int label = 0;
fprintf(txtfile, "%d,", label); //use "," instead of "\t"
}
float f_testdata[DIM_OF_SAMPLE96X32] = { 0.0f };
for (int i = 0; i < DIM_OF_SAMPLE96X32; i++)
{
if (i < DIM_OF_HOG96X32)
{
f_testdata[i] = descriptors0[i];
fprintf(txtfile, "%f,", descriptors0[i]);
}
else if (i<DIM_OF_HOG96X32+DIM_OF_LBP1)
{
f_testdata[i] = LBPH00.at<float>(0, i - DIM_OF_HOG96X32);
fprintf(txtfile, "%f,", LBPH00.at<float>(0, i - DIM_OF_HOG96X32));
}
else if (i == DIM_OF_SAMPLE96X32-2)
{
f_testdata[i] = (float)src.rows*src.cols / 1000.0f;
fprintf(txtfile, "%f,", (float)src.rows*src.cols / 1000.0f);
}
else
{
f_testdata[i] = max((float)src.rows / (float)src.cols, (float)src.cols / (float)src.rows);
fprintf(txtfile, "%f\n", max((float)src.rows / (float)src.cols, (float)src.cols / (float)src.rows));
}
}
//negative sample features...
}
//train negative end...
}
else
{
//测试样本
for (int img_ind_pos = 0; img_ind_pos != 384; img_ind_pos++)
{
sprintf(srcimgs, "/home/jumper/Ecology_projects/Algaes_train_test/zhilianzao/zhilianzao_fortest/%d.JPG", img_ind_pos);
Mat src = imread(srcimgs,0);
if(!src.data)
{
continue;
}
cout<<"extract feature of image---"<<img_ind_pos<<"..."<<endl;
//...此处代码不能显示
//生成测试样本的特征
for (int i = 0; i < DIM_OF_SAMPLE96X32; i++)
{
if (i < DIM_OF_HOG96X32)
{
f_testdata[i] = descriptors0[i];
fprintf(txtfile, "%f,", descriptors0[i]);
}
else if (i<DIM_OF_HOG96X32+DIM_OF_LBP1)
{
f_testdata[i] = LBPH00.at<float>(0, i - DIM_OF_HOG96X32);
fprintf(txtfile, "%f,", LBPH00.at<float>(0, i - DIM_OF_HOG96X32));
}
else if (i == DIM_OF_SAMPLE96X32-2)
{
f_testdata[i] = (float)src.rows*src.cols / 1000.0f;
fprintf(txtfile, "%f,", (float)src.rows*src.cols / 1000.0f);
}
else
{
f_testdata[i] = max((float)src.rows / (float)src.cols, (float)src.cols / (float)src.rows);
fprintf(txtfile, "%f\n", max((float)src.rows / (float)src.cols, (float)src.cols / (float)src.rows));
}
}
//features...
}
//test end...
}
fclose(txtfile);
return 0;
}然后再在xgboost下训练测试:
import numpy as np
import pandas as pd
import xgboost as xgb
from sklearn.cross_validation import train_test_split
import os,glob
import re
from PIL import Image
numbers=re.compile(r'(\d+)')
def numericalSort(value):
parts=numbers.split(value)
parts[1:2]=map(int,parts[1::2])
return parts
#记录程序运行时间
import time
start_time = time.time()
#读入样本特征数据(libsvm格式或者numpy格式,我的是numpy格式)
train = pd.read_csv("/home/jumper/Ecology_projects/Algaes_train_test/zhilianzao/train/train.csv")
train_sz = train.shape
train_xy,val = train_test_split(train, test_size = 0.1,random_state=1)
y = train_xy.Label
X = train_xy.drop(["Label"],axis=1)
val_y = val.Label
val_X = val.drop(["Label"],axis=1)
xgb_val = xgb.DMatrix(val_X,label=val_y)
xgb_train = xgb.DMatrix(X, label=y)
params={
'booster':'gbtree',
'objective': 'binary:logistic', #二分类
#'num_class':5, # 多分类的问题 类别数,与 'objective': 'multi:softmax '并用
'gamma':0.6, # 用于控制是否后剪枝的参数,越大越保守,一般0.1、0.2这样子。
'max_depth':9, # 构建树的深度,越大越容易过拟合
'lambda':2, # 控制模型复杂度的权重值的L2正则化项参数,参数越大,模型越不容易过拟合。
'subsample':0.7, # 随机采样训练样本
'colsample_bytree':0.9, # 生成树时进行的列采样
'min_child_weight':3,
# 这个参数默认是 1,是每个叶子里面 h 的和至少是多少,对正负样本不均衡时的 0-1 分类而言
#,假设 h 在 0.01 附近,min_child_weight 为 1 意味着叶子节点中最少需要包含 100 个样本。
#这个参数非常影响结果,控制叶子节点中二阶导的和的最小值,该参数值越小,越容易 overfitting。
'silent':0 ,#设置成1则没有运行信息输出,最好是设置为0.
'eta': 0.1, # 如同学习率
'seed':1000,
'nthread':5,# cpu 线程数
'eval_metric': 'error',
'reg_alpha':10
#'updater'= 'grow_gpu'
}
plst = list(params.items())
num_rounds = 150 # 迭代次数
watchlist = [(xgb_train, 'train'),(xgb_val, 'val')]
#训练模型并保存
# early_stopping_rounds 当设置的迭代次数较大时,early_stopping_rounds 可在一定的迭代次数内准确率没有提升就停止训练
mymodel = xgb.train(plst, xgb_train, num_rounds, watchlist,early_stopping_rounds=100)
mymodel.save_model('/home/jumper/Ecology_projects/Algaes_train_test/zhilianzao/zhilianzao_xgb.model') # 用于存储训练出的模型
print ("best best_ntree_limit",mymodel.best_ntree_limit )
#输出运行时长
cost_time = time.time()-start_time
print ("xgboost success!",'\n',"cost time:",cost_time,"(s)......")
###########################################################################
##########################################################################
#########################################################################
imgs_pos_folder="/home/jumper/Ecology_projects/Algaes_train_test/zhilianzao/zhilianzao_fortest"
imgsextension2=['JPG']
os.chdir(imgs_pos_folder)
imglist_pos=[]
for extension in imgsextension2:
extension='*.'+extension
imglist_pos+=[os.path.realpath(e) for e in glob.glob(extension)]
pos_imgslist=sorted(imglist_pos,key=numericalSort)
tests = pd.read_csv("/home/jumper/Ecology_projects/Algaes_train_test/zhilianzao/zhilianzao_fortest/test.csv")
xgb_test = xgb.DMatrix(tests)
###########################################################################
##########################################################################
#########################################################################
#unknown test sets...use my new model!
#save images to different folders according to predict-classes..
preds = mymodel.predict(xgb_test,ntree_limit=mymodel.best_ntree_limit)
#predict classes possibility...
#np.savetxt('/home/jumper/Ecology_projects/Algaes_train_test/zhilianzao/test/xgb_submission.csv',np.c_[range(1,len(tests)+1),preds],delimiter=',',header='ImageId,Label',comments='',fmt='%d')
#predict classes...
preds_classes=np.arange(1,len(preds)+1)
for i in range(0,len(preds)):
if preds[i]>0.5:
preds_classes[i]=1
theimg=Image.open(pos_imgslist[i])
grayimg=theimg.convert('L')
grayimg.save("/home/jumper/Ecology_projects/Algaes_train_test/zhilianzao/predict/positive/"+str(i)+".jpg")
else:
preds_classes[i]=0
theimg=Image.open(pos_imgslist[i])
grayimg=theimg.convert('L')
grayimg.save("/home/jumper/Ecology_projects/Algaes_train_test/zhilianzao/predict/negative/"+str(i)+".jpg")
np.savetxt('/home/jumper/Ecology_projects/Algaes_train_test/zhilianzao/test/xgb_result.csv',np.c_[range(1,len(tests)+1),preds_classes],delimiter=',',header='ImageId,Label',comments='',fmt='%d') 0x00007ffff34926f2 in std::_Rb_tree<int, std::pair<int const, std::__cxx11::basic_string<char, std::char_traits<char>, std::allocator<char> > >, std::_Select1st<std::pair<int const, std::__cxx11::basic_string<char, std::char_traits<char>, std::allocator<char> > > >, std::less<int>, std::allocator<std::pair<int const, std::__cxx11::basic_string<char, std::char_traits<char>, std::allocator<char> > > > >::_M_erase(std::_Rb_tree_node<std::pair<int const, std::__cxx11::basic_string<char, std::char_traits<char>, std::allocator<char> > > >*) ()
from /home/jumper/workspace/xgboostV0.7/lib/libxgboost.so”
我又换成了我原来的XGBoost版本(C++11下的),不再出现此问题。估计是他编译的这个库是未启用c++11特性则其中的std::string实际上是std::basic_string<char> ,而默认情况下,GCC 5在编译时会将std::string类型按c++11下std::__cxx11::basic_string<char> 来处理。解决这个问题/类似问题的根本方法就是在编译时不要混用c++11和c++03库/代码。
/****************************************************************************************************************/
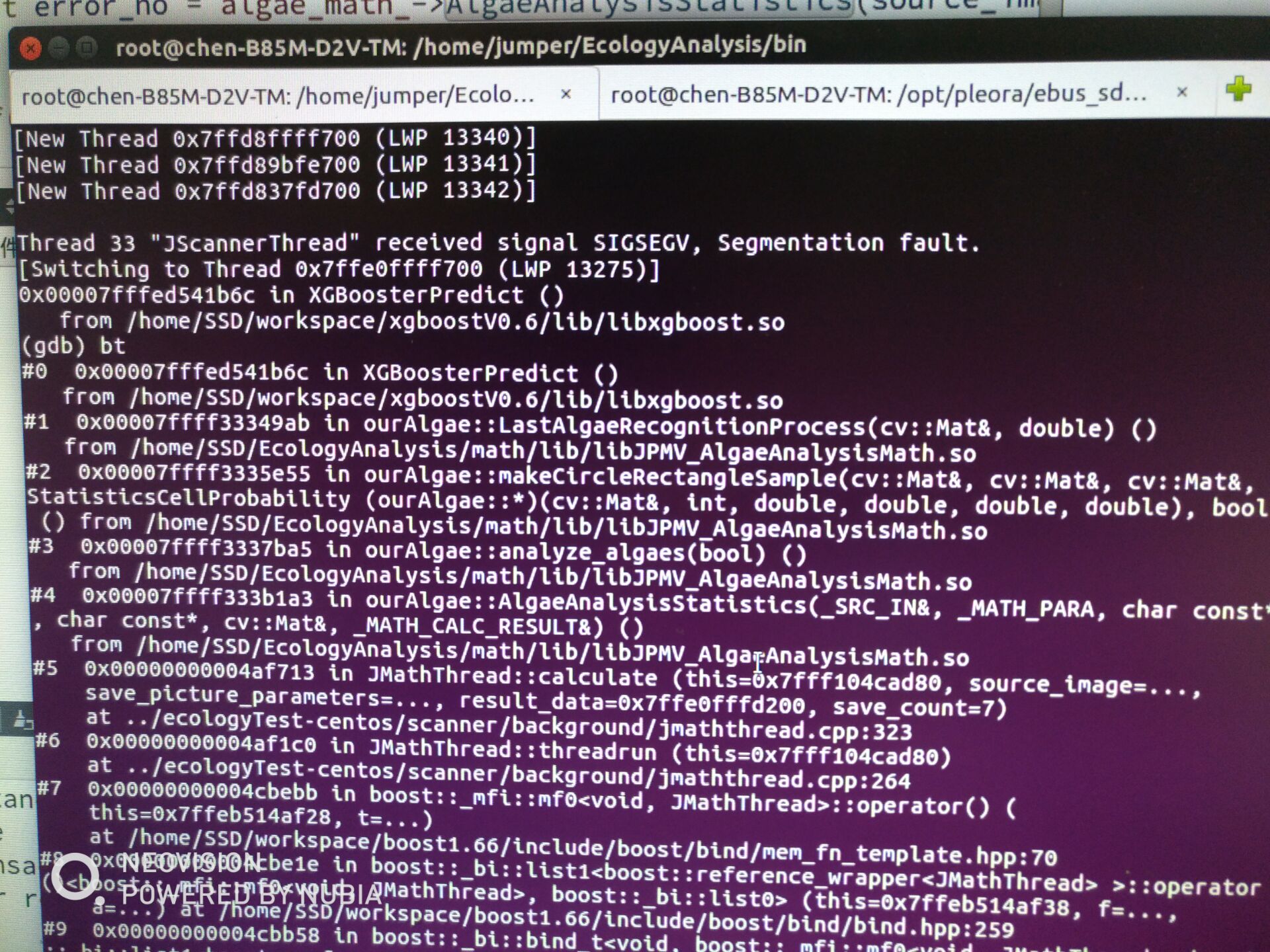
今天2018.5.28,遇到一个问题:Ubuntu下XGBoost多线程下调用XGBoosterPredict()崩溃显示:0x00007fffed541b6c in XGBoosterPredict(),如下图:
ubuntu单线程下却没有问题;同样的代码CentOS下单线程多线程都没问题 这个问题还在查找中。。。?
五、多线程的另一个例子(利用线程池)
例子的src部分我放在:http://download.csdn.net/download/wd1603926823/9926757
本来是开10个线程运行到第两百多张图时 终端显示“已杀死”!这个是指针操作导致哪里的内存问题,但我检查了几遍仍旧觉得指针操作无误,很无奈所以将指针全部换掉了就好了!
六、多线程(利用线程池)单例、锁
多了几个库需要被用到:boost_filesystem boost_serialization
多线程单例加锁:http://download.csdn.net/download/wd1603926823/9935747
多线程单例不加锁:http://download.csdn.net/download/wd1603926823/9935987
多个线程对同一块内存写时才需要加锁(加锁会影响性能)

























 1752
1752











 被折叠的 条评论
为什么被折叠?
被折叠的 条评论
为什么被折叠?










