参考链接:
http://blog.csdn.net/u010902721/article/details/23531359
http://blog.csdn.net/czrzchao/article/details/52314455
本例遗传算法求
最大值
代码如下:
import random
import math
import numpy as np
import matplotlib.pyplot as plt
population_size = 500 # 种群数量
generations = 200 # 迭代次数
chrom_length = 10 # 染色体长度
pc = 0.6 # 交配概率
pm = 0.01 # 变异概率
genetic_population = [] # 种群的基因编码(二进制)
population = [] # 种群对应的十进制数值,并标准化范围到[0, 10]
fitness = [] # 适应度
fitness_mean = []
optimum_solution = [] # 每次迭代的所获得的最优解
# 为染色体进行0,1编码,生成初始种群
def chrom_encoding():
for i in range(population_size):
population_i = []
for j in range(chrom_length):
population_i.append(random.randint(0, 1))
genetic_population.append(population_i)
# 对染色体进行解码,将二进制转化为十进制
def chrom_decoding():
population.clear()
for i in range(population_size):
value = 0
for j in range(chrom_length):
value += genetic_population[i][j] * (2 ** (chrom_length - 1 - j))
population.append(value * 10 / (2 ** (chrom_length) - 1))
# 计算每个染色体的适应度
def calculate_fitness():
sum = 0.0
fitness.clear()
for i in range(population_size):
function_value = 10 * math.sin(5 * population[i]) + 7 * math.cos(4 * population[i])
if function_value > 0.0:
sum += function_value
fitness.append(function_value)
else:
fitness.append(0.0)
# 返回群体的平均适应度
return sum / population_size
# 获取最大适应度的个体和对应的编号
def best_value():
max_fitness = fitness[0]
max_chrom = 0
for i in range(population_size):
if fitness[i] > max_fitness:
max_fitness = fitness[i]
max_chrom = i
return max_chrom, max_fitness
# 采用轮盘赌算法进行选择过程,重新选择与种群数量相等的新种群
def selection():
fitness_proportion = []
fitness_sum = 0
for i in range(population_size):
fitness_sum += fitness[i]
# 计算生存率
for i in range(population_size):
fitness_proportion.append(fitness[i] / fitness_sum)
pie_fitness = []
cumsum = 0.0
for i in range(population_size):
pie_fitness.append(cumsum + fitness_proportion[i])
cumsum += fitness_proportion[i]
pie_fitness[-1] = 1
# 生成随机数在轮盘上选点[0, 1)
random_selection = []
for i in range(population_size):
random_selection.append(random.random())
random_selection.sort()
# 选择新种群
new_genetic_population = []
random_selection_id = 0
global genetic_population
for i in range(population_size):
while random_selection_id < population_size and random_selection[random_selection_id] < pie_fitness[i]:
new_genetic_population.append(genetic_population[i])
random_selection_id += 1
genetic_population = new_genetic_population
'''
# 用numpy的random.choice函数直接模拟轮盘赌算法
def selection():
fitness_array = np.array(fitness)
new_population_id = np.random.choice(np.arange(population_size), (population_size,),
replace=True, p=fitness_array/fitness_array.sum())
new_genetic_population = []
global genetic_population
for i in range(population_size):
new_genetic_population.append(genetic_population[new_population_id[i]])
genetic_population = new_genetic_population
'''
# 进行交配过程
def crossover():
for i in range(0, population_size - 1, 2):
if random.random() < pc:
# 随机选择交叉点
change_point = random.randint(0, chrom_length - 1)
temp1 = []
temp2 = []
temp1.extend(genetic_population[i][0: change_point])
temp1.extend(genetic_population[i+1][change_point:])
temp2.extend(genetic_population[i+1][0: change_point])
temp2.extend(genetic_population[i][change_point:])
genetic_population[i] = temp1
genetic_population[i+1] = temp2
# 进行基因的变异
def mutation():
for i in range(population_size):
if random.random() < pm:
mutation_point = random.randint(0, chrom_length - 1)
if genetic_population[i][mutation_point] == 0:
genetic_population[i][mutation_point] = 1
else:
genetic_population[i][mutation_point] = 0
chrom_encoding()
for step in range(generations):
chrom_decoding()
fit_mean = calculate_fitness()
best_id, best_fitness = best_value()
optimum_solution.append(best_fitness)
fitness_mean.append(fit_mean)
selection()
crossover()
mutation()
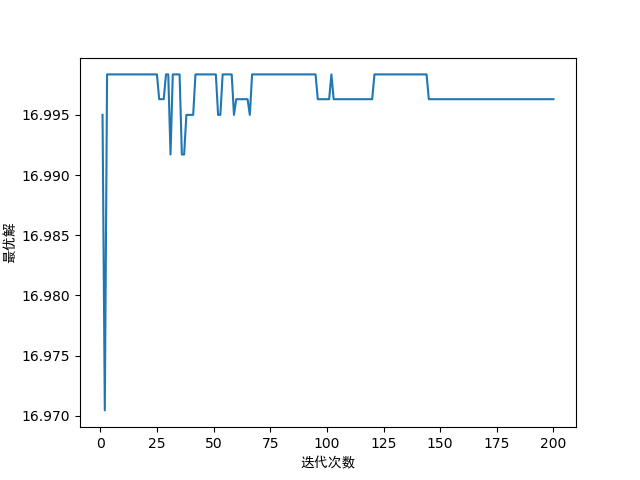
# 最优解随迭代次数的变化
fig1 = plt.figure(1)
plt.plot(range(1, generations + 1), optimum_solution)
plt.xlabel('迭代次数', fontproperties='SimHei')
plt.ylabel('最优解', fontproperties='SimHei')
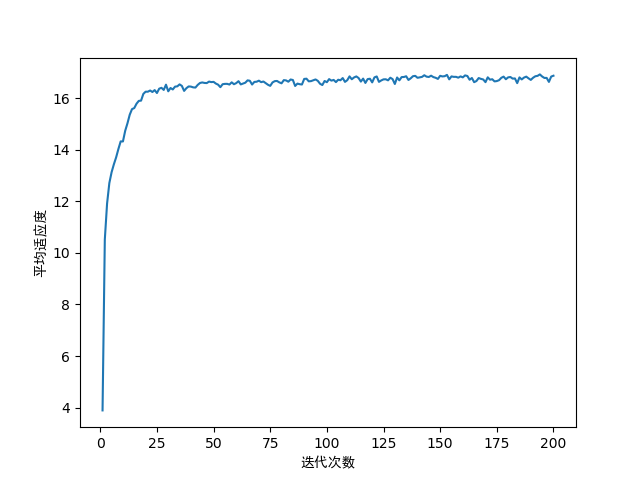
# 平均适应度随迭代次数的变化
fig2 = plt.figure(2)
plt.plot(range(1, generations + 1), fitness_mean)
plt.xlabel('迭代次数', fontproperties='SimHei')
plt.ylabel('平均适应度', fontproperties='SimHei')
plt.show()
最优解与迭代次数的关系如下:
平均适应度与迭代次数的关系:
利用微生物遗传算法(Microbial GA)解决上述最值问题
该算法主要是对传统遗传算法进行改进
算法思想:对于选择过程,在population中,随机选择2组DNA,比较2者fitness,大的fitness者为winner,小的fitness者为loser,保持winner不变,只对loser进行改变。loser在与winner交叉过程中大约以pc的部分获得winner的DNA,然后再对loser进行变异,最后将处理后的2组DNA放入population。
本算法的好处是可以保留当代的优良种群到下一代,而且必须是产生比当代最优良个体还优良的个体才能把当前优良个体取代,使得在迭代过程中best fitness只会增加不会下降
import numpy as np
import random
import matplotlib.pyplot as plt
population_size = 50
pc = 0.6
pm = 0.01
n_generations = 500
DNA_length = 10
x_bound = [0, 10]
fig = plt.figure()
def F(x):
return 10 * np.sin(5 * x) + 7 * np.cos(4 * x)
class GA(object):
def __init__(self):
self.populations = np.random.randint(0, 2, (population_size, DNA_length))
# 对所有DNA进行解码,翻译成10进制数字
def DNA_decode(self, loser_winner):
return np.dot(loser_winner, 2 ** np.arange(DNA_length)) / (2 ** DNA_length-1) * x_bound[1]
def calculate_fitness(self, loser_winner):
DNA_value = self.DNA_decode(loser_winner)
fitness = F(DNA_value)
fitness = np.where(fitness < 0.0, 0.0, fitness)
return fitness
def selection(self):
# 从0~DNA.length-1中随机选择2组DNA编号
loser_winner_id = np.random.choice(np.arange(population_size), size=2, replace=False)
loser_winner = self.populations[loser_winner_id]
loser_winner_fitness = self.calculate_fitness(loser_winner)
return loser_winner[np.argsort(loser_winner_fitness)], loser_winner_id
def crossover(self, loser_winner):
# 必须指定类型为np.bool,否则True会变为1.0000的小数
crossover_points = np.empty((DNA_length,)).astype(np.bool)
for i in range(DNA_length):
crossover_points[i] = True if random.random() < pc else False
loser_winner[0, crossover_points] = loser_winner[1, crossover_points]
return loser_winner
def mutation(self, loser_winner):
for i in range(DNA_length):
loser_winner[0, i] = 1 if loser_winner[0, i] == 0 else 0
return loser_winner
def evolve(self):
for i in range(int(population_size/2)):
loser_winner, loser_winner_id = self.selection()
loser_winner = self.crossover(loser_winner)
loser_winner = self.mutation(loser_winner)
self.populations[loser_winner_id] = loser_winner
population_value = np.linspace(x_bound[0], x_bound[1], 200)
plt.plot(population_value, F(population_value))
ga = GA()
for step in range(n_generations):
population_fitness = ga.calculate_fitness(ga.populations)
x = ga.DNA_decode(ga.populations[np.argmax(population_fitness)])
y_max = np.max(population_fitness)
y_mean = np.mean(population_fitness)
# 描点
# globals以字典类型返回当前位置的全部全局变量
if 'sca' in globals():
sca.remove()
sca = plt.scatter(x, y_max, s=100, c='red')
plt.pause(0.01)
print('the best fitness: %.3f, the mean fitness: %.3f' % (y_max, y_mean))
ga.evolve()
plt.show()
























 77
77

 被折叠的 条评论
为什么被折叠?
被折叠的 条评论
为什么被折叠?








