1.题目解读
数据集采用经典的鸢尾花分类问题数据集如下:
| 序号 | 花萼长度 | 花萼宽度 | 花瓣长度 | 花瓣宽度 | 种类 |
| 1 | 5.1 | 3.5 | 1.4 | 0.2 | setosa |
| 2 | 4.9 | 3 | 1.4 | 0.2 | setosa |
| 3 | 4.7 | 3.2 | 1.3 | 0.2 | setosa |
| 4 | 4.6 | 3.1 | 1.5 | 0.2 | setosa |
| 5 | 5 | 3.6 | 1.4 | 0.2 | setosa |
| 6 | 5.4 | 3.9 | 1.7 | 0.4 | setosa |
| 7 | 4.6 | 3.4 | 1.4 | 0.3 | setosa |
| 8 | 5 | 3.4 | 1.5 | 0.2 | setosa |
| 9 | 4.4 | 2.9 | 1.4 | 0.2 | setosa |
| 10 | 4.9 | 3.1 | 1.5 | 0.1 | setosa |
| 11 | 5.4 | 3.7 | 1.5 | 0.2 | setosa |
| 12 | 4.8 | 3.4 | 1.6 | 0.2 | setosa |
| 13 | 4.8 | 3 | 1.4 | 0.1 | setosa |
| 14 | 4.3 | 3 | 1.1 | 0.1 | setosa |
| 15 | 5.8 | 4 | 1.2 | 0.2 | setosa |
| 16 | 5.7 | 4.4 | 1.5 | 0.4 | setosa |
| 17 | 5.4 | 3.9 | 1.3 | 0.4 | setosa |
| 18 | 5.1 | 3.5 | 1.4 | 0.3 | setosa |
| 19 | 5.7 | 3.8 | 1.7 | 0.3 | setosa |
| 20 | 5.1 | 3.8 | 1.5 | 0.3 | setosa |
| 21 | 5.4 | 3.4 | 1.7 | 0.2 | setosa |
| 22 | 5.1 | 3.7 | 1.5 | 0.4 | setosa |
| 23 | 4.6 | 3.6 | 1 | 0.2 | setosa |
| 24 | 5.1 | 3.3 | 1.7 | 0.5 | setosa |
| 25 | 4.8 | 3.4 | 1.9 | 0.2 | setosa |
| 26 | 5 | 3 | 1.6 | 0.2 | setosa |
| 27 | 5 | 3.4 | 1.6 | 0.4 | setosa |
| 28 | 5.2 | 3.5 | 1.5 | 0.2 | setosa |
| 29 | 5.2 | 3.4 | 1.4 | 0.2 | setosa |
| 30 | 4.7 | 3.2 | 1.6 | 0.2 | setosa |
| 31 | 4.8 | 3.1 | 1.6 | 0.2 | setosa |
| 32 | 5.4 | 3.4 | 1.5 | 0.4 | setosa |
| 33 | 5.2 | 4.1 | 1.5 | 0.1 | setosa |
| 34 | 5.5 | 4.2 | 1.4 | 0.2 | setosa |
| 35 | 4.9 | 3.1 | 1.5 | 0.2 | setosa |
| 36 | 5 | 3.2 | 1.2 | 0.2 | setosa |
| 37 | 5.5 | 3.5 | 1.3 | 0.2 | setosa |
| 38 | 4.9 | 3.6 | 1.4 | 0.1 | setosa |
| 39 | 4.4 | 3 | 1.3 | 0.2 | setosa |
| 40 | 5.1 | 3.4 | 1.5 | 0.2 | setosa |
| 41 | 5 | 3.5 | 1.3 | 0.3 | setosa |
| 42 | 4.5 | 2.3 | 1.3 | 0.3 | setosa |
| 43 | 4.4 | 3.2 | 1.3 | 0.2 | setosa |
| 44 | 5 | 3.5 | 1.6 | 0.6 | setosa |
| 45 | 5.1 | 3.8 | 1.9 | 0.4 | setosa |
| 46 | 4.8 | 3 | 1.4 | 0.3 | setosa |
| 47 | 5.1 | 3.8 | 1.6 | 0.2 | setosa |
| 48 | 4.6 | 3.2 | 1.4 | 0.2 | setosa |
| 49 | 5.3 | 3.7 | 1.5 | 0.2 | setosa |
| 50 | 5 | 3.3 | 1.4 | 0.2 | setosa |
| 51 | 7 | 3.2 | 4.7 | 1.4 | versicolor |
| 52 | 6.4 | 3.2 | 4.5 | 1.5 | versicolor |
| 53 | 6.9 | 3.1 | 4.9 | 1.5 | versicolor |
| 54 | 5.5 | 2.3 | 4 | 1.3 | versicolor |
| 55 | 6.5 | 2.8 | 4.6 | 1.5 | versicolor |
| 56 | 5.7 | 2.8 | 4.5 | 1.3 | versicolor |
| 57 | 6.3 | 3.3 | 4.7 | 1.6 | versicolor |
| 58 | 4.9 | 2.4 | 3.3 | 1 | versicolor |
| 59 | 6.6 | 2.9 | 4.6 | 1.3 | versicolor |
| 60 | 5.2 | 2.7 | 3.9 | 1.4 | versicolor |
| 61 | 5 | 2 | 3.5 | 1 | versicolor |
| 62 | 5.9 | 3 | 4.2 | 1.5 | versicolor |
| 63 | 6 | 2.2 | 4 | 1 | versicolor |
| 64 | 6.1 | 2.9 | 4.7 | 1.4 | versicolor |
| 65 | 5.6 | 2.9 | 3.6 | 1.3 | versicolor |
| 66 | 6.7 | 3.1 | 4.4 | 1.4 | versicolor |
| 67 | 5.6 | 3 | 4.5 | 1.5 | versicolor |
| 68 | 5.8 | 2.7 | 4.1 | 1 | versicolor |
| 69 | 6.2 | 2.2 | 4.5 | 1.5 | versicolor |
| 70 | 5.6 | 2.5 | 3.9 | 1.1 | versicolor |
| 71 | 5.9 | 3.2 | 4.8 | 1.8 | versicolor |
| 72 | 6.1 | 2.8 | 4 | 1.3 | versicolor |
| 73 | 6.3 | 2.5 | 4.9 | 1.5 | versicolor |
| 74 | 6.1 | 2.8 | 4.7 | 1.2 | versicolor |
| 75 | 6.4 | 2.9 | 4.3 | 1.3 | versicolor |
| 76 | 6.6 | 3 | 4.4 | 1.4 | versicolor |
| 77 | 6.8 | 2.8 | 4.8 | 1.4 | versicolor |
| 78 | 6.7 | 3 | 5 | 1.7 | versicolor |
| 79 | 6 | 2.9 | 4.5 | 1.5 | versicolor |
| 80 | 5.7 | 2.6 | 3.5 | 1 | versicolor |
| 81 | 5.5 | 2.4 | 3.8 | 1.1 | versicolor |
| 82 | 5.5 | 2.4 | 3.7 | 1 | versicolor |
| 83 | 5.8 | 2.7 | 3.9 | 1.2 | versicolor |
| 84 | 6 | 2.7 | 5.1 | 1.6 | versicolor |
| 85 | 5.4 | 3 | 4.5 | 1.5 | versicolor |
| 86 | 6 | 3.4 | 4.5 | 1.6 | versicolor |
| 87 | 6.7 | 3.1 | 4.7 | 1.5 | versicolor |
| 88 | 6.3 | 2.3 | 4.4 | 1.3 | versicolor |
| 89 | 5.6 | 3 | 4.1 | 1.3 | versicolor |
| 90 | 5.5 | 2.5 | 4 | 1.3 | versicolor |
| 91 | 5.5 | 2.6 | 4.4 | 1.2 | versicolor |
| 92 | 6.1 | 3 | 4.6 | 1.4 | versicolor |
| 93 | 5.8 | 2.6 | 4 | 1.2 | versicolor |
| 94 | 5 | 2.3 | 3.3 | 1 | versicolor |
| 95 | 5.6 | 2.7 | 4.2 | 1.3 | versicolor |
| 96 | 5.7 | 3 | 4.2 | 1.2 | versicolor |
| 97 | 5.7 | 2.9 | 4.2 | 1.3 | versicolor |
| 98 | 6.2 | 2.9 | 4.3 | 1.3 | versicolor |
| 99 | 5.1 | 2.5 | 3 | 1.1 | versicolor |
| 100 | 5.7 | 2.8 | 4.1 | 1.3 | versicolor |
| 101 | 6.3 | 3.3 | 6 | 2.5 | virginica |
| 102 | 5.8 | 2.7 | 5.1 | 1.9 | virginica |
| 103 | 7.1 | 3 | 5.9 | 2.1 | virginica |
| 104 | 6.3 | 2.9 | 5.6 | 1.8 | virginica |
| 105 | 6.5 | 3 | 5.8 | 2.2 | virginica |
| 106 | 7.6 | 3 | 6.6 | 2.1 | virginica |
| 107 | 4.9 | 2.5 | 4.5 | 1.7 | virginica |
| 108 | 7.3 | 2.9 | 6.3 | 1.8 | virginica |
| 109 | 6.7 | 2.5 | 5.8 | 1.8 | virginica |
| 110 | 7.2 | 3.6 | 6.1 | 2.5 | virginica |
| 111 | 6.5 | 3.2 | 5.1 | 2 | virginica |
| 112 | 6.4 | 2.7 | 5.3 | 1.9 | virginica |
| 113 | 6.8 | 3 | 5.5 | 2.1 | virginica |
| 114 | 5.7 | 2.5 | 5 | 2 | virginica |
| 115 | 5.8 | 2.8 | 5.1 | 2.4 | virginica |
| 116 | 6.4 | 3.2 | 5.3 | 2.3 | virginica |
| 117 | 6.5 | 3 | 5.5 | 1.8 | virginica |
| 118 | 7.7 | 3.8 | 6.7 | 2.2 | virginica |
| 119 | 7.7 | 2.6 | 6.9 | 2.3 | virginica |
| 120 | 6 | 2.2 | 5 | 1.5 | virginica |
| 121 | 6.9 | 3.2 | 5.7 | 2.3 | virginica |
| 122 | 5.6 | 2.8 | 4.9 | 2 | virginica |
| 123 | 7.7 | 2.8 | 6.7 | 2 | virginica |
| 124 | 6.3 | 2.7 | 4.9 | 1.8 | virginica |
| 125 | 6.7 | 3.3 | 5.7 | 2.1 | virginica |
| 126 | 7.2 | 3.2 | 6 | 1.8 | virginica |
| 127 | 6.2 | 2.8 | 4.8 | 1.8 | virginica |
| 128 | 6.1 | 3 | 4.9 | 1.8 | virginica |
| 129 | 6.4 | 2.8 | 5.6 | 2.1 | virginica |
| 130 | 7.2 | 3 | 5.8 | 1.6 | virginica |
| 131 | 7.4 | 2.8 | 6.1 | 1.9 | virginica |
| 132 | 7.9 | 3.8 | 6.4 | 2 | virginica |
| 133 | 6.4 | 2.8 | 5.6 | 2.2 | virginica |
| 134 | 6.3 | 2.8 | 5.1 | 1.5 | virginica |
| 135 | 6.1 | 2.6 | 5.6 | 1.4 | virginica |
| 136 | 7.7 | 3 | 6.1 | 2.3 | virginica |
| 137 | 6.3 | 3.4 | 5.6 | 2.4 | virginica |
| 138 | 6.4 | 3.1 | 5.5 | 1.8 | virginica |
| 139 | 6 | 3 | 4.8 | 1.8 | virginica |
| 140 | 6.9 | 3.1 | 5.4 | 2.1 | virginica |
| 141 | 6.7 | 3.1 | 5.6 | 2.4 | virginica |
| 142 | 6.9 | 3.1 | 5.1 | 2.3 | virginica |
| 143 | 5.8 | 2.7 | 5.1 | 1.9 | virginica |
| 144 | 6.8 | 3.2 | 5.9 | 2.3 | virginica |
| 145 | 6.7 | 3.3 | 5.7 | 2.5 | virginica |
| 146 | 6.7 | 3 | 5.2 | 2.3 | virginica |
| 147 | 6.3 | 2.5 | 5 | 1.9 | virginica |
| 148 | 6.5 | 3 | 5.2 | 2 | virginica |
| 149 | 6.2 | 3.4 | 5.4 | 2.3 | virginica |
| 150 | 5.9 | 3 | 5.1 | 1.8 | virginica |
2.思路分析
由于特征比较少(仅有4个),所有没有采取随机森林的标准方法,而是使用了bagging方法如下图。

其中先对于每一个基学习器,训练集通过自助采样法在原始数据集中进行采样获得,并将剩余数据作为测试集。(约37.2%)
基学习器算法采用基于ID3的决策树算法,具体如下图

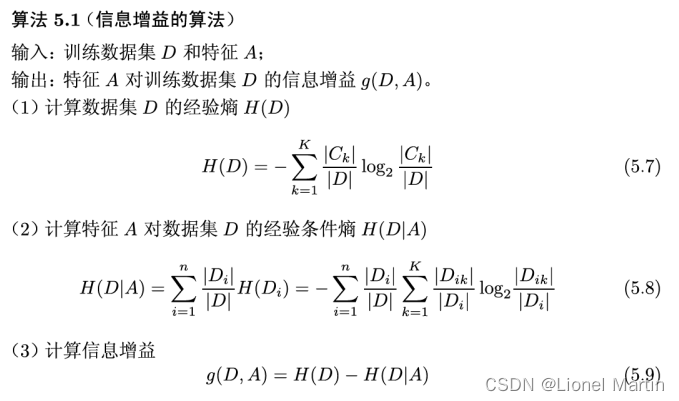
其中信息增益率为
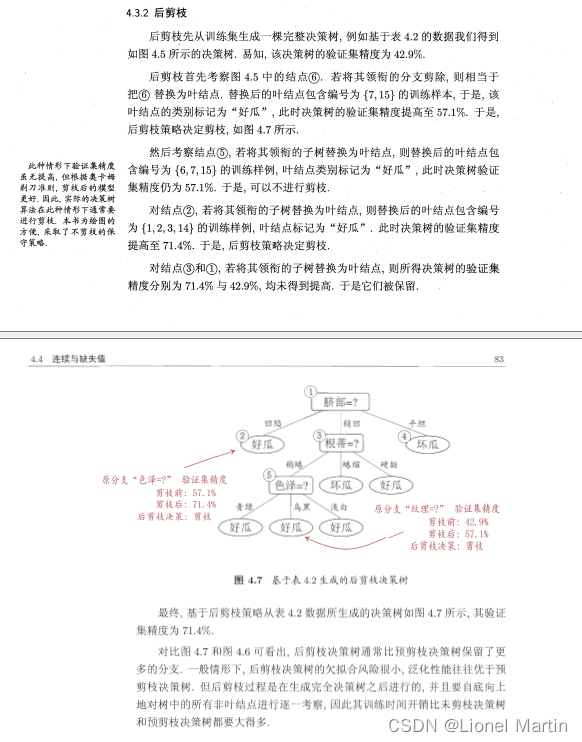
对决策树的剪枝采取后剪枝操作,具体如下:
3.代码实现
主要由以下几个函数构成:
-
全局变量设置:
NameSet:包含数据集中所有类别的集合。Name:类别名称与数字标识的映射。trans:数字标识与类别名称的映射。
-
数据读取与预处理:
- 使用
pandas库读取CSV文件。 - 对数据进行标准化处理,以便于后续的计算。
- 使用
-
决策树构建函数:
calculate:计算给定数据集的最佳信息增益比。CreateTree:根据数据集创建决策树。check_part:递归检查决策树的每个节点。crosscheck:验证决策树的准确性。Simplify:简化决策树,进行剪枝。GetNodeNum:获取决策树的节点数。GetClassfier:获取分类器和验证集。NoPruning:创建决策树但不进行剪枝。left和right:获取决策树的左子树和右子树。seg_veridata:根据决策树分割验证数据集。Pruning:对决策树进行剪枝。
-
随机森林(Bagging)实现:
Ensemble:集成多个分类器的预测结果。ErrorEstimate:估计分类器的错误率。bagging和bagging_nopruning:分别实现带剪枝和不带剪枝的随机森林。BaseLearner:生成一系列基学习器并计算它们的准确率。
-
参数设置与调整:
SetParameter:设置决策树和随机森林的参数。SetRate:调整剪枝参数并计算准确率。
-
绘图与结果保存:
draw_base:绘制基学习器数量与准确率的关系图。draw_rate:绘制剪枝参数与准确率的关系图。draw_prun:比较剪枝与不剪枝的准确率差异。- 使用
matplotlib库进行绘图,并保存图像。 - 使用
pandas库将准确率数据保存为Excel文件。
-
代码执行与测试:
- 代码中包含了对上述函数的测试和示例执行。
- 通过调整参数和观察绘图结果来评估模型性能。
具体代码如下:
import pandas as pd
import numpy as np
import math
import matplotlib.pyplot as plt
import random
import string
from matplotlib import pyplot as plt
plt.rcParams['font.sans-serif'] = ['SimHei'] # 中文字体设置-黑体
plt.rcParams['axes.unicode_minus'] = False # 解决保存图像是负号'-'显示为方块的问题
#全局变量设置
NameSet = {"setosa","versicolor","virginica"}
Name = {1:"setosa",2:"versicolor",3:"virginica"}
trans = {"setosa":1,"versicolor":2,"virginica":3}
#print(Name[1])
#读入数据
data = pd.read_csv("iris2.csv",header=None)
data = np.array(data)
data_copy = data
#print(data_copy)
num = len(data[:,5])
#对数据进行标准化处理
#参数设置
setrate = 0.2
bound = 0
T = 100
testnum = 10
PruningCheck = 1
for i in range(0,len(data[:,5])):
data[i][5] = trans[ data[i][5] ]
for i in range(1,5):
Max = data[:,i].max()
Min = data[:,i].min()
for j in range(0,len(data[:,i])):
data[j][i] = (Max - data[j][i]) / (Max - Min)
#给定数据集计算最佳信息增益比
def seg_data(seq,data):
#print(seq)
out = np.zeros((len(seq),6))
for i in range(0,len(seq)):
out[i,:] = data[int(seq[i]),:]
return out
def calculate(data):
if len(data[:,5]) <= 1:
return "NULL"
#计算信息增益比
num = len(data[:,5])
#计算H(D)
cnt = [0]*4
for i in range(0,num):
data[i][5] = int(data[i][5])
for i in range(0,num):
cnt[ int(data[i][5]) ] += 1
H_D = 0
for i in range(1,4):
if cnt[i] != 0:
H_D += -(cnt[i]/num) * math.log2(cnt[i]/num)
def Gain(j,t,data):
num = len(data[:,5])
stat = np.zeros((3,4))
for i in range(0,num):
if data[i][j] < t:
tex = 1
else:
tex = 2
stat[tex][ int(data[i][5]) ] += 1
#print(stat)
H_DA = 0
for k in range(1,3):
tmp = 0
for i in range(1,4):
if stat[k][i] != 0:
tmp += -stat[k][i]/num * math.log2( stat[k][i]/num )
tmp *= np.sum(stat[k,:])/num
#print(np.sum(stat[k,:]))
H_DA += tmp
return H_D-H_DA
GAIN = np.zeros(5)
T = np.zeros(5)
for i in range(1,5):
#print(data[:,i])
t_seq = np.sort(data[:,i])
#print(t_seq)
t_copy = [0] * ( len(t_seq) - 1 )
for j in range(0,len(t_copy)):
t_copy[j] = ( t_seq[j] + t_seq[j+1] ) / 2
#print(t_copy)
gain = [0] * len(t_copy)
for j in range(0,len( t_copy )):
gain[j] = Gain(i,t_copy[j],data)
#print(t_copy[j], gain[j])
#if np.size(gain) != 0:
#if np.size(gain) < 10:
# print(data)
"""
print(np.size(gain) )
print(t_copy)
print(gain)
print(data)
"""
tmp = np.argmax(gain)
GAIN[i] = gain[tmp]
T[i] = t_copy[tmp]
seg_cate = np.argmax(T)
seg_value = np.max(T)
info_increase = np.max(GAIN)
seq_low = []
seq_high = []
for i in range(0,num):
if data[i][seg_cate] < seg_value:
seq_low += [i]
else:
seq_high += [i]
data_low = seg_data(seq_low,data)
data_high = seg_data(seq_high,data)
cnt = np.zeros(4)
for i in range(0,len(data)):
cnt[ int(data[i][5]) ] += 1
most_cate = np.argmax(cnt)
output = (seg_cate,seg_value,info_increase,data_low,data_high,most_cate)
return output
#print(GAIN,T)
#定义信息增益阈值
#bound = 0
def CreateTree(data):
#检测是否类标签相同
for i in range(1,len(data)):
if data[i][5] != data[i-1][5]:
break
if i == len(data) - 1:
return Name[ data[i][5] ]
seg = calculate(data)
#print(seg[0:3],seg[5])
#print( np.size(seg[3]) , np.size(seg[4]) )
if seg == "NULL":
return Name[data[0][5]]
if np.size(seg[3]) * np.size(seg[4]) == 0:
return Name [ seg[5] ]
if seg[2] < bound:
return Name[ seg[5] ]
mytree = { seg[0] :{round(seg[1],2):{ "fir":CreateTree(seg[3]),"sec":CreateTree(seg[4])} } }
return mytree
#mytree = CreateTree(data)
#print(mytree)
def check_part(mytree,test):
#print(mytree)
if type(mytree) == type("setosa"):
return mytree
key = list( mytree.keys() )
cate = key[0]
cate_key = list ( mytree[ cate ] .keys() )
seg = cate_key[0]
#print(test,seg)
if test[ cate ] < seg:
mytree = mytree[ cate ][ seg ][ 'fir' ]
else:
mytree = mytree[ cate ][ seg ][ 'sec' ]
#print(type(mytree))
if type(mytree) == type("str"):
return mytree
return check_part ( mytree,test )
def crosscheck(mytree,data):
#print(mytree)
#print(data)
num = len(data[:,5])
#print("num=",num)
cnt = 0
for i in range(0,num):
res = check_part(mytree,data[i,:])
#print(res,data[i][5])
if res == Name[ int(data[i][5]) ]:
cnt += 1
return cnt
#设置基于Bagging的训练轮数T
#T = 50
def Simplify(mytree):
if type(mytree) == type("setosa"):
return mytree
key = list( mytree.keys() )
cate = key[0]
cate_key = list ( mytree[ cate ] .keys() )
seg = cate_key[0]
if type(mytree[ cate ][ seg ][ 'fir' ]) != type("setosa"):
mytree[ cate ][ seg ][ 'fir'] = Simplify( mytree[ cate ][ seg ][ 'fir'] )
if type(mytree[ cate ][ seg ][ 'sec' ]) != type("setosa"):
mytree[ cate ][ seg ][ 'sec' ] = Simplify( mytree[ cate ][ seg ][ 'sec'] )
if type(mytree[ cate ][ seg ][ 'fir' ]) == type("setosa") and type(mytree[ cate ][ seg ][ 'sec' ]) == type("setosa") and mytree[ cate ][ seg ][ 'fir' ] == mytree[ cate ][ seg ][ 'sec' ]:
mytree = mytree[ cate ][ seg ][ 'fir' ]
return mytree
return mytree
def GetNodeNum(mytree):
if type(mytree) == type("setosa"):
return 0
key = list( mytree.keys() )
cate = key[0]
cate_key = list ( mytree[ cate ] .keys() )
seg = cate_key[0]
cnt = 1
#if type(mytree[ cate ][ seg ][ 'fir' ]) != type("setosa"):
cnt += GetNodeNum( mytree[ cate ][ seg ][ 'fir'] )
#if type(mytree[ cate ][ seg ][ 'sec' ]) != type("setosa"):
cnt += GetNodeNum( mytree[ cate ][ seg ][ 'sec'] )
return cnt
def GetClassfier(data):
num = len(data[:,5])
data_seq = np.zeros(num)
for i in range(0,num):
tmp = random.randint(0,num-1)
data_seq[i] = tmp
tmp = []
for i in range(0,num):
if i not in data_seq:
tmp.append(i)
veri_seq = np.array(tmp)
dataset = seg_data(data_seq,data)
#print(len(tmp))
veriset = seg_data(veri_seq,data)
#print("veriset=",veriset)
mytree = CreateTree(dataset)
#print("old=",mytree)
mytree = Simplify(mytree)
if PruningCheck == 1:
mytree = Pruning(mytree,veriset)
#print("nodenum=",GetNodeNum(mytree),"mytree=",mytree)
return (mytree,veri_seq)
def NoPruning(data):
num = len(data[:,5])
data_seq = np.zeros(num)
for i in range(0,num):
tmp = random.randint(0,num-1)
data_seq[i] = tmp
tmp = []
for i in range(0,num):
if i not in data_seq:
tmp.append(i)
veri_seq = np.array(tmp)
dataset = seg_data(data_seq,data)
#print(len(tmp))
veriset = seg_data(veri_seq,data)
#print("veriset=",veriset)
mytree = CreateTree(dataset)
#print("old=",mytree)
mytree = Simplify(mytree)
#mytree = Pruning(mytree,veriset)
#print("nodenum=",GetNodeNum(mytree),"mytree=",mytree)
return (mytree,veri_seq)
def left(mytree):
key = list( mytree.keys() )
cate = key[0]
cate_key = list ( mytree[ cate ] .keys() )
seg = cate_key[0]
return mytree[ cate ][ seg ][ 'fir']
def right(mytree):
key = list( mytree.keys() )
cate = key[0]
cate_key = list ( mytree[ cate ] .keys() )
seg = cate_key[0]
return mytree[ cate ][ seg ][ 'sec']
def seg_veridata(mytree,data):
key = list( mytree.keys() )
cate = key[0]
cate_key = list ( mytree[ cate ] .keys() )
seg = cate_key[0]
leftseq = []
rightseq = []
num = len(data[:,5])
for i in range(0,num):
if data[ i ][ cate ] < seg:
leftseq += [i]
else:
rightseq += [i]
leftdata = seg_data(leftseq,data)
rightdata = seg_data(rightseq,data)
return (leftdata,rightdata)
def Pruning(mytree,data):
num = len(data[:,5])
if type(mytree) == type("str") or num == 0:
return mytree
key = list( mytree.keys() )
cate = key[0]
cate_key = list ( mytree[ cate ] .keys() )
seg = cate_key[0]
res = seg_veridata(mytree, data)
leftdata = res[0]
rightdata = res[1]
mytree[ cate ][ seg ][ 'fir'] = Pruning( left(mytree) , leftdata)
mytree[ cate ][ seg ][ 'sec'] = Pruning( right(mytree) , rightdata)
unpruning_rate = crosscheck(mytree,data)/num
cnt = np.zeros(4)
for i in range(0,num):
#print(data[i][5])
cnt[ int(data[i][5]) ] += 1
pruning_rate = np.max(cnt)/num
# print(cnt,"num=",num,"mytree=",mytree)
pruning_name = Name[ np.argmax(cnt) ]
if pruning_rate - unpruning_rate > setrate:
mytree = pruning_name
return mytree
def Ensemble(classfier,data_setlist,test):
cnt = np.zeros(4)
for i in range(0,T):
if test[0] not in data_setlist[i]:
#print( check_part( classfier[i] , test ) )
cnt[ trans[ check_part( classfier[i] , test ) ] ]+= 1
print(cnt)
return np.argmax(cnt)
def ErrorEstimate(classfier,data_setlist,data):
num = len(data[:,5])
cnt = 0
for i in range(0,num):
if Ensemble(classfier,data_setlist, data[i,:] ) == data[i][5]:
cnt += 1
return cnt/num
def bagging(data):
classfier = [0] * T
data_setlist = [0] * T
for i in range(0,T):
res = GetClassfier(data)
#print(i)
classfier[i] = res[0]
data_setlist[i] = res[1]
#print(mytree)
#print("check")
error = ErrorEstimate(classfier,data_setlist,data)
return error
#bagging(data)
def bagging_nopruning(data):
classfier = [0] * T
data_setlist = [0] * T
for i in range(0,T):
res = NoPruning(data)
#print(i)
classfier[i] = res[0]
data_setlist[i] = res[1]
#print(mytree)
#print("check")
error = ErrorEstimate(classfier,data_setlist,data)
return error
nopruning = np.zeros(10)
pruning = np.zeros(10)
"""
for i in range(0,testnum):
nopruning[i] = bagging_nopruning(data)
pruning[i] = bagging(data)
print("nopruning=",nopruning[i],"pruning=",pruning[i],"delta=",pruning[i]-nopruning[i])
print("avg_delta=",(np.sum(pruning)-np.sum(nopruning))/testnum)
"""
setrate = 0
bound = 0
T = 100
testnum = 10
PruningCheck = 1
Parameter = [0.2,0,100,10,1]
def SetParameter(Parameter):
global setrate
global bound
global T
global testnum
global PruningCheck
setrate = Parameter[0]
bound = Parameter[1]
T = Parameter[2]
testsnum = Parameter[3]
PruningCheck = Parameter[4]
Ques = [
[0,5.9,2.6,3.8,1.2],
[0,6.1,2.6,5.3,1.7],
[0,5.1,3.5,1.4,0.2],
[0,7.2,3.6,6.1,2.5]
]
tmp = pd.read_csv("iris2.csv",header=None)
data_copy = np.array(tmp)
#print(data_copy)
for i in range(0,len(Ques)):
for j in range(1,5):
#print(data_copy)
max_data = np.max(data_copy[:,j])
min_data = np.min(data_copy[:,j])
#print(data_copy[:,j])
#print(max_data,min_data)
Ques[i][j] = (max_data - Ques[i][j]) / (max_data - min_data)
#print(Ques)
classifier = [0] * T
for i in range(0,T):
res = GetClassfier(data)
classifier[i] = res[0]
dataset = []
for i in range(0,T):
dataset.append([])
#print(dataset)
for i in range(0,len(Ques)):
print( "第"+str(i+1)+"支花的类别是:",Name[ Ensemble(classifier, dataset, Ques[i] ) ] )
def PerformanceAnalysis(data):
Parameter = [0.2,0,10,10,1]
SetParameter(Parameter)
Total_T = 10
def BaseLearner(Total_T):
bagging_data = np.zeros(Total_T)
for i in range(1,Total_T + 1):
print(i)
Parameter[2] = i
SetParameter(Parameter)
#print("T=",T)
bagging_data[i-1] = bagging(data)
#print(i,bagging_data[i-1])
return bagging_data
bagging_data = BaseLearner(Total_T)
x=range(1,Total_T + 1)
#设置y
y=bagging_data
#plot函数需要两个参数,一个是x一个是y
def draw_base(x,y):
plt.plot(x,y,"ob:")
plt.title('基学习器数量和准确率的关系图')
plt.xlabel('基学习器数量')
plt.ylabel('准确率')
plt.legend()
draw_base(x,y)
plt.show()
draw_base(x,y)
plt.savefig('基学习器数量对准确率的影响.png')
op = pd.DataFrame(y)
#op.to_excel('y.xlsx')
Parameter = [0.2,0,10,10,1]
def SetRate(maxrate):
bagging_data = np.zeros(int(maxrate/min_step + 1))
y_unpruning = np.zeros(int(maxrate/min_step + 1))
for i in np.arange(0,maxrate+min_step,min_step):
Parameter[0] = i
PruningCheck = 1
SetParameter(Parameter)
#print("setrate=",setrate)
bagging_data[ int(i/min_step) ] = bagging(data)
PruningCheck = 0
y_unpruning[ int(i/min_step) ] = bagging(data)
print(i,bagging_data[ int(i/min_step) ],y_unpruning[ int(i/min_step) ])
#print("setrate=",setrate,bagging_data[ int(i/min_step) ])
return (bagging_data,y_unpruning)
def draw_rate(x,y,y_unpruning):
#plt.plot(x,y,"ob:")
plt.plot(x,y,"ob:",label="pruning")
plt.plot(x,y_unpruning,"or:",label="unpruning")
plt.title('剪枝参数和准确率的关系图')
plt.xlabel('剪枝参数')
plt.ylabel('准确率')
plt.legend()
min_step = 0.02
maxrate = 0.1
res = SetRate(maxrate)
x = np.arange(0,maxrate+min_step,min_step)
#print(x)
#print(np.size(x))
#设置y
y = res[0]
y_unpruning = res[1]
draw_rate(x,y,y_unpruning)
plt.show()
draw_rate(x,y,y_unpruning)
plt.savefig('剪枝参数对准确率的影响.png')
out = np.zeros((2, np.size(y) ))
out[0,:] = y
out[1,:] = y_unpruning
op = pd.DataFrame(out)
#op.to_excel('pruning or not.xlsx')
#PerformanceAnalysis(data)
"""
"""
"""
Parameter = [0.2,0,10,10,1]
SetParameter(Parameter)
num_set = 5
x = range(1,num_set + 1)
y_pruning = np.zeros(num_set)
y_unpruning = np.zeros(num_set)
for i in range(0,num_set):
PruningCheck = 1
y_pruning[i] = bagging(data)
PruningCheck = 0
y_unpruning[i] = bagging(data)
print("剪枝平均单次误差提升",( np.sum(y_pruning)-np.sum(y_unpruning) ) / num_set)
def draw_prun():
plt.plot(x,y_pruning,"ob:",label="pruning")
plt.plot(x,y_unpruning,"or:",label="unpruning")
plt.title('剪枝参数和准确率的关系图')
plt.xlabel('次数')
plt.ylabel('准确率')
plt.legend()
draw_prun()
plt.show()
draw_prun()
plt.savefig('是否剪枝对准确率的影响.png')
#create_plot(mytree)
#op = pd.DataFrame(data)
#op.to_excel('data1.xlsx')
"""
#基于ID3算法实现决策树构建
"""
"""
{3: {0.75: {'fir': {2: {0.81: {'fir': {2: {0.73: {'fir': {2: {0.69: {'fir': {2: {0.6: {'fir': {1: {0.54: {'fir': {1: {0.49: {'fir': {3: {0.31: {'fir': 'virginica',
'sec': 'versicolor'}}},
'sec': 'versicolor'}}},
'sec': 'versicolor'}}},
'sec': {2: {0.65: {'fir': 'versicolor',
'sec': 'virginica'}}}}}},
'sec': 'versicolor'}}},
'sec': {2: {0.77: {'fir': 'versicolor', 'sec': 'versicolor'}}}}}},
'sec': 'versicolor'}}},
'sec': 'setosa'}}}
"""
"""
4.结果分析
主要通过由以下几个部分构成:
1. 评估基学习器数量对于泛化能力的提升
2. 评估剪枝参数和是否剪枝对于泛化能力的提升
这里先对几个重要参数做说明:
Setrate( )表示剪枝容忍度,即对于用于包外剪枝的数据集,如果将一棵树直接换成判断节点比原来的树的准确度好Setrate时就决定剪枝。
T 表示基学习器数量,即使用几个基学习器参与投票进行分类
PruningCheck 表示是否剪枝,0代表不剪枝,1代表剪枝。
通过运行程序,我们可以获得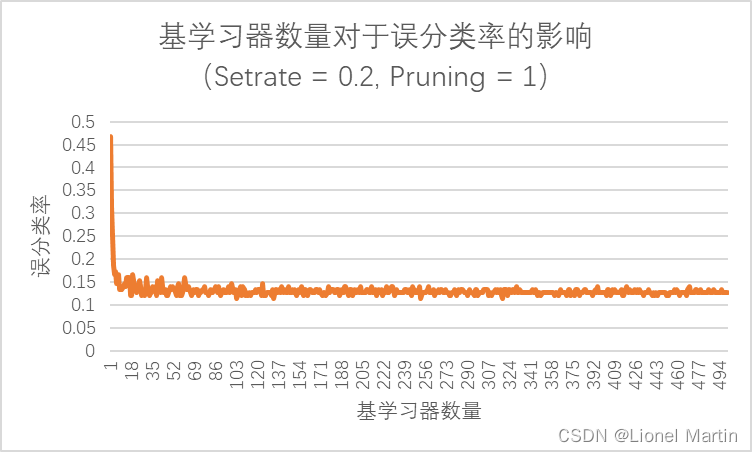
观察图片可知bagging集成技术对于误分类率的优化是现象级的,当T=1时,ER (ErrorRate) = 46.67%. 但是当T=20时,ER = 14%. 但是当基学习器大幅度增加时,误分类率未见明显增长基本稳定在13%左右,即
分类准确率 = 87%
分析剪枝参数对于准确率的影响我们有以下图片: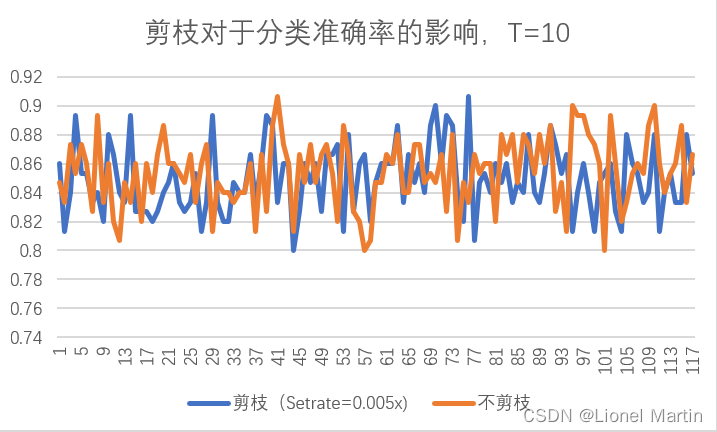
其中Setrate从0按步长0.005逐渐变化到0.6
剪枝平均准确率 0.834958
不剪枝平均准确率 0.846779
相对差值 -0.01182
分析可知,平均来说,剪枝准确率甚至比不剪枝的平均准确率还少了约1%,但结合图像变化,可以认为,剪枝操作对准确率的提升不大。
5.后记
制作这个项目零零散散花费了一周的时间,希望下次再代码编写上能过更加熟练一点




 博客围绕鸢尾花分类问题展开,采用bagging方法和基于ID3的决策树算法。介绍了代码实现,包括数据读取、决策树构建、随机森林实现等函数。结果分析评估了基学习器数量和剪枝参数对泛化能力的影响,发现bagging能优化误分类率,剪枝对准确率提升不大。
博客围绕鸢尾花分类问题展开,采用bagging方法和基于ID3的决策树算法。介绍了代码实现,包括数据读取、决策树构建、随机森林实现等函数。结果分析评估了基学习器数量和剪枝参数对泛化能力的影响,发现bagging能优化误分类率,剪枝对准确率提升不大。

















 736
736

 被折叠的 条评论
为什么被折叠?
被折叠的 条评论
为什么被折叠?








