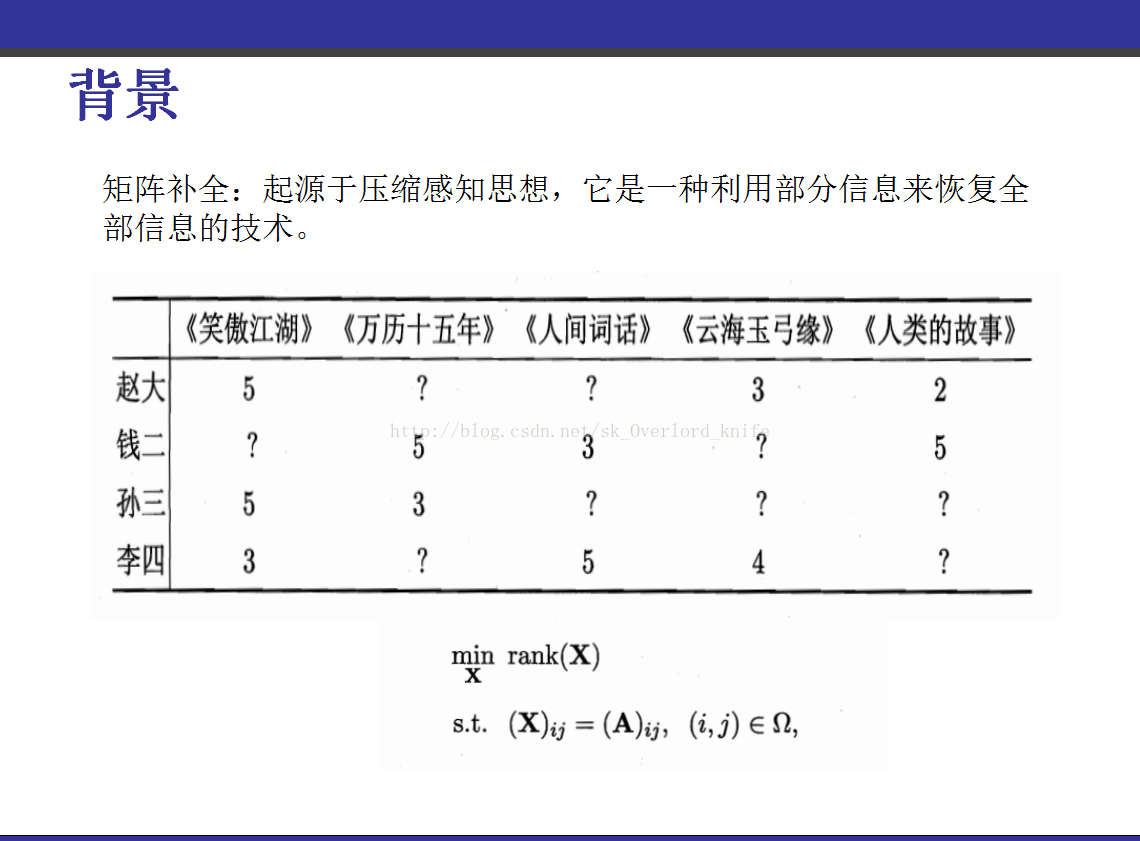
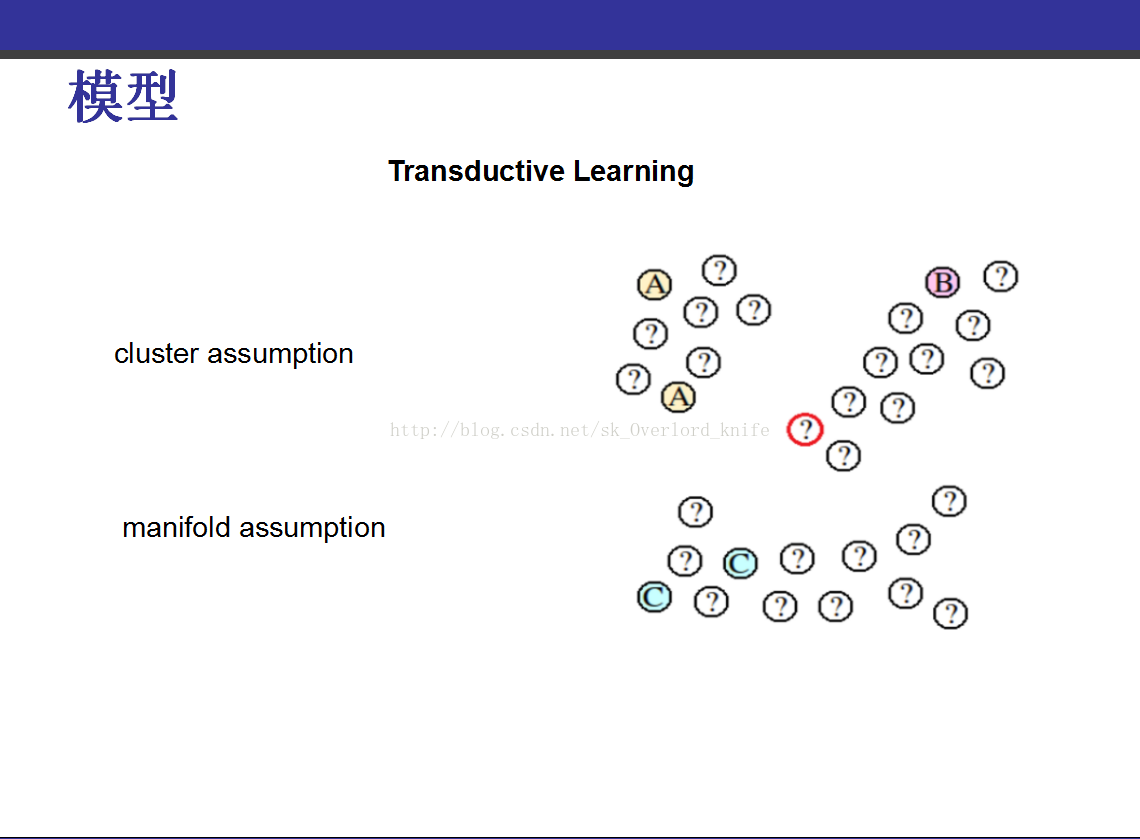
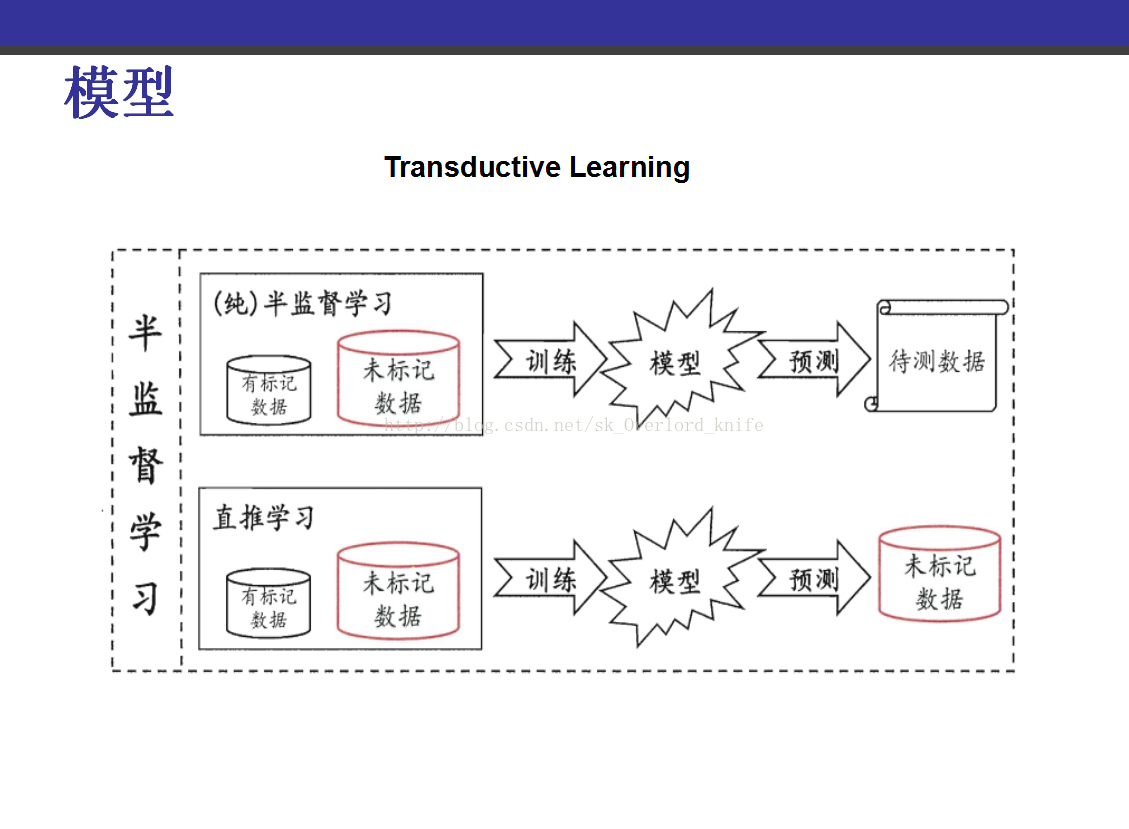
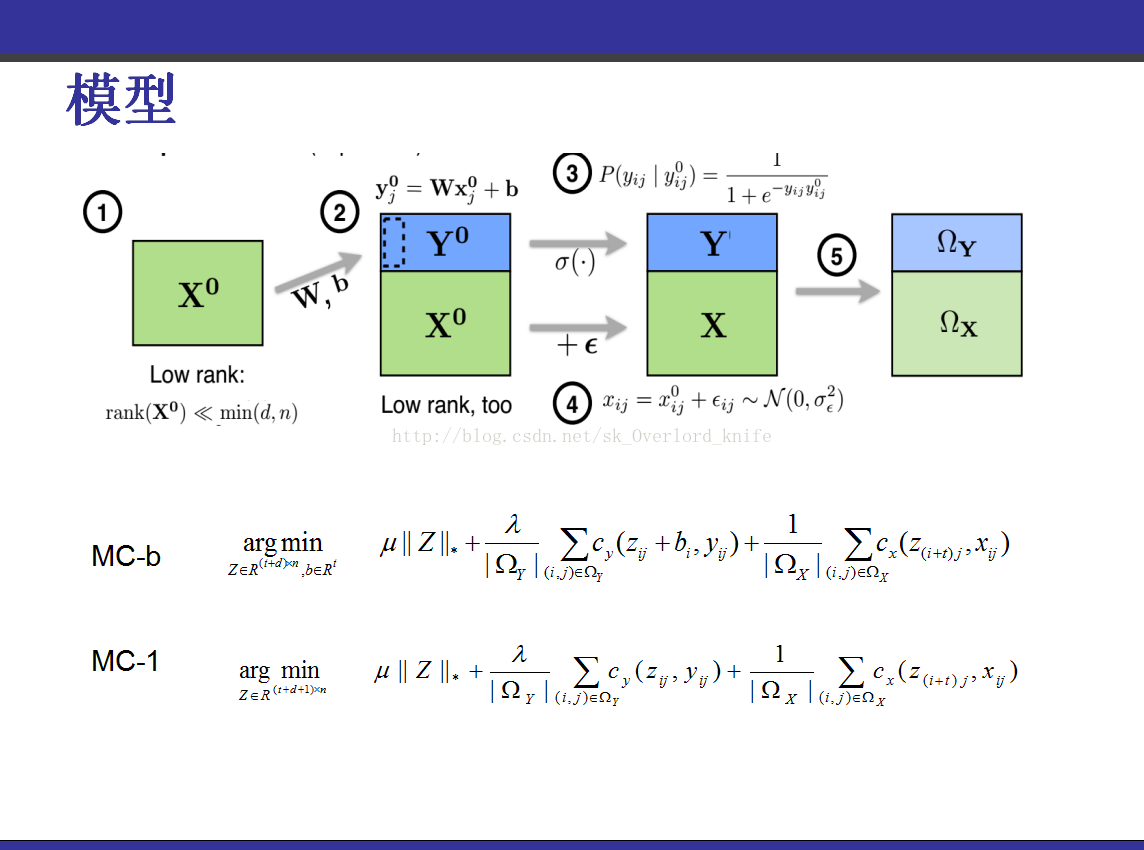
该PPT讲解的是《transduction-with-matrix-completion-three-birds-with-one-stone》,这篇论文
参考文献:
基于前向后向算子分裂的稀疏信号重构-
Fixed-point continuation for l1-minimization methodology and convergence
Convergence of fixed point continuation algorithms for matrix rank minimization
Fixed point and Bregman iterative methods for matrix rank minimization
补充知识:次微分
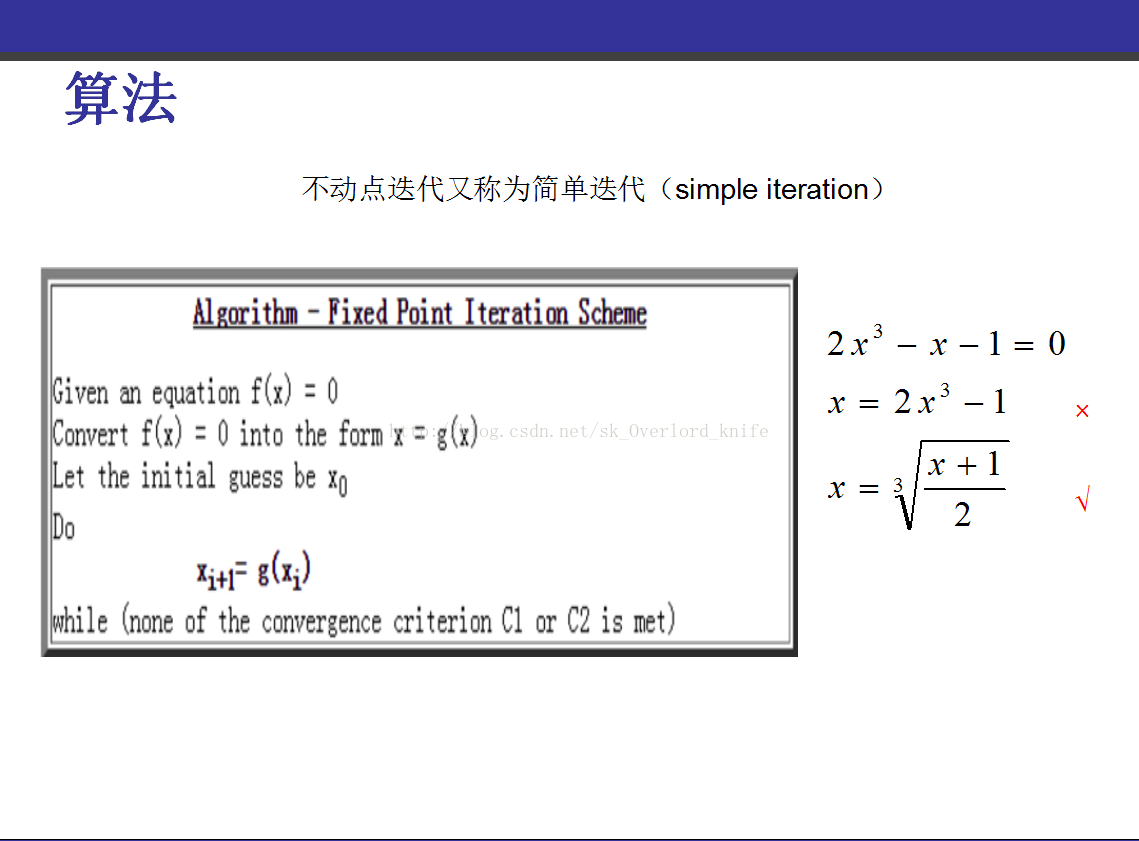
不动点迭代 http://blog.csdn.net/jbb0523/article/details/52459797?reload
求解线性方程组的迭代法(直接搜百度找知识点)
不动点迭代可以参考 Numerical Analysis (美)Timothy Sauer 这本书
参考代码:
Test_MC_1.m
%% test script
% initialize matrix
clear;
clc;
add_bias = 0;
if ispc
% reorth.f isn't compiled for Windows, but this shouldn't be an
% issue, because the .m file version is pretty fast
warning('off','PROPACK:NotUsingMex');
end
%% Setup a matrix
for s1 = 1:1
randn('state',2009+s1);
rand('state',2009+s1);
% if you're daring, try with complex numbers:
COMPLEX = false;
NOISE_LEVEL= 0.0;
n1 = 400; n2 = 180; r = 50;
if COMPLEX
X = (randn(n1,r)+1i*randn(n1,r))*(randn(r,n2)+1i*randn(r,n2))/2;
else
X = randn(n1,r)*randn(r,n2)+rand(n1,n2)*NOISE_LEVEL;
end
W = rand(n2,1);
Y = sign(X*W-mean(X*W)+rand(n1,1)*NOISE_LEVEL);
% add in noise, if desired
sigma = 0;
% sigma = .05*std(data);
% data = data + sigma*randn(size(data));
df = r*(n1+n2-r);
oversampling = 5;
m = min(5*df,round(.90*n1*n2) );
p = m/(n1*n2);
Omega{1} = sort(randsample(n1*n2,m)); % Omega = randperm(n1*n2); Omega = Omega(1:m);
Omega{2} = sort(randsample(n1,round(p*n1)));
data{1} = X(Omega{1});
data{2} = Y(Omega{2});
% % --- adding bias term (all ones to X)----
% X = [X ones(n1,1)];
% Omega{1} = [sort(randsample(n1*n2,m)) ; (n1*n2+(1:n1))'];
% data{1} = X(Omega{1});
% % -----------------------------
missingX = ones(n1,n2);
missingY = ones(n1,1);
missingX(Omega{1}) = 0;
missingY(Omega{2}) = 0;
if add_bias == 1
missingX((n1*(n2-1)+1):(n1*n2)) = 0;
end
missingX = logical(missingX);
missingY = logical(missingY);
fprintf('Matrix completion: %d x %d matrix, rank %d, %.1f%% observations\n',...
n1,n2,r,100*p);
fprintf('\toversampling degrees of freedom by %.1f; noise std is %.1e\n',...
m/df, NOISE_LEVEL );
string1 = sprintf('Matrix completion: %d x %d matrix, rank %d, %.1f%% observations\n',...
n1,n2,r,100*p);
string2 = sprintf('\toversampling degrees of freedom by %.1f; noise std is %.1e\n',...
m/df, NOISE_LEVEL );
if ~isreal(X), disp('Matrix is complex'); end
% Set parameters and solve
tau = 5*sqrt(n1*n2);
delta = 1.2/p;
maxiter = 500;
tol = 1e-4;
% Approximate minimum nuclear norm solution by FPC algorithm
% This version of FPC uses PROPACK for the multiplies
% It is not optimized, and the parameters have not been tested
% The version in the FPC paper by Shiqian Ma, Donald Goldfarb and Lifeng Chen
% uses an approximate SVD that will have different properties;
% That code may be found at http://www.columbia.edu/~sm2756/FPCA.htm
tol = 1e-3;
% lambda_set = [0.005 0.5 5 10 20 50 100 200 500 1000];
lambda_set = [0.0005 0.005 0.05 0.5 1 5];
% mu_final_set = [0.000001 0.00001 0.0001] ;
mu_final_set = [0.000001 0.00001 0.0001 0.001 ] ;
fprintf('\nSolving by FPC...\n');
for i = 1:length(lambda_set)
for j = 1:length(mu_final_set)
lambda = lambda_set(i);
mu_final = mu_final_set(j);
[U,S,V,numiter] = TMC([n1 n2,1],Omega,data,mu_final,lambda,maxiter,tol);
Z = U*S*V';
% OmegaAll = [Omega{1} ;n1*(n2)+Omega{2}]; % if FPC_kh1 is used
OmegaAll = [Omega{1} ;n1*(n2+1)+Omega{2}]; % if FPC_kh2 is used
Yp = Z(:,end);
Xp = Z(:,1:end-1);
Yp_miss = Yp(missingY); % missingY is mask of missing data, Omega is location of known data
Xp_miss = Xp(missingX);
% accuracy on Y prediction
% [acc(j,i,s1), sen(j,i,s1), spe(j,i,s1)] = AccSenSpe(Yp_miss,Y(missingY));
acc(j,i) = sum(sign(Yp_miss) == Y(missingY))/length(Yp_miss);
re_X(j,i,s1) = norm(Xp_miss - X(missingX),'fro')/norm(X(missingX),'fro');
re_whole(i,j) = norm([X ones(n1,1) Y]-Z,'fro')/norm([X ones(n1,1) Y],'fro'); % if FPC_kh2 is used
% re_whole(j,i,s1) = norm([X Y]-Z,'fro')/norm([X Y],'fro'); % if FPC_kh1 is used
% Show results
fprintf('The recovered rank is %d\n',length(diag(S)) );
fprintf('The relative error on Omega is: %d\n', norm([data{1}; data{2}]-Z(OmegaAll))/norm([data{1}; data{2}]))
% fprintf('The relative recovery error is: %d\n', norm([X ones(n1,1) Y]-Z,'fro')/norm([X ones(n1,1) Y],'fro'))
% fprintf('The relative recovery in the spectral norm is: %d\n', norm([X Y]-Z)/norm([X Y]))
fprintf('The relative recovery error on missing X is: %.4f\n', re_X(j,i));
fprintf('Output Y prediction accuracy is: %.3f\n', acc(j,i));
result.rank(j,i,s1) = length(diag(S));
% result.acc = acc;
% result.sen = sen;
% result.spe = spe;
result.re_X = re_X;
result.re_whole = re_whole;
result.info = [string1 string2];
end
end
end
% save result_random_test3_FPC_kh1 result
% save result_random_test1_FPC_kh2_extrabreak result
[mean(re_X,3) ;mean(re_Y,3) mean(result.rank,3)]
======================================================================================================================================
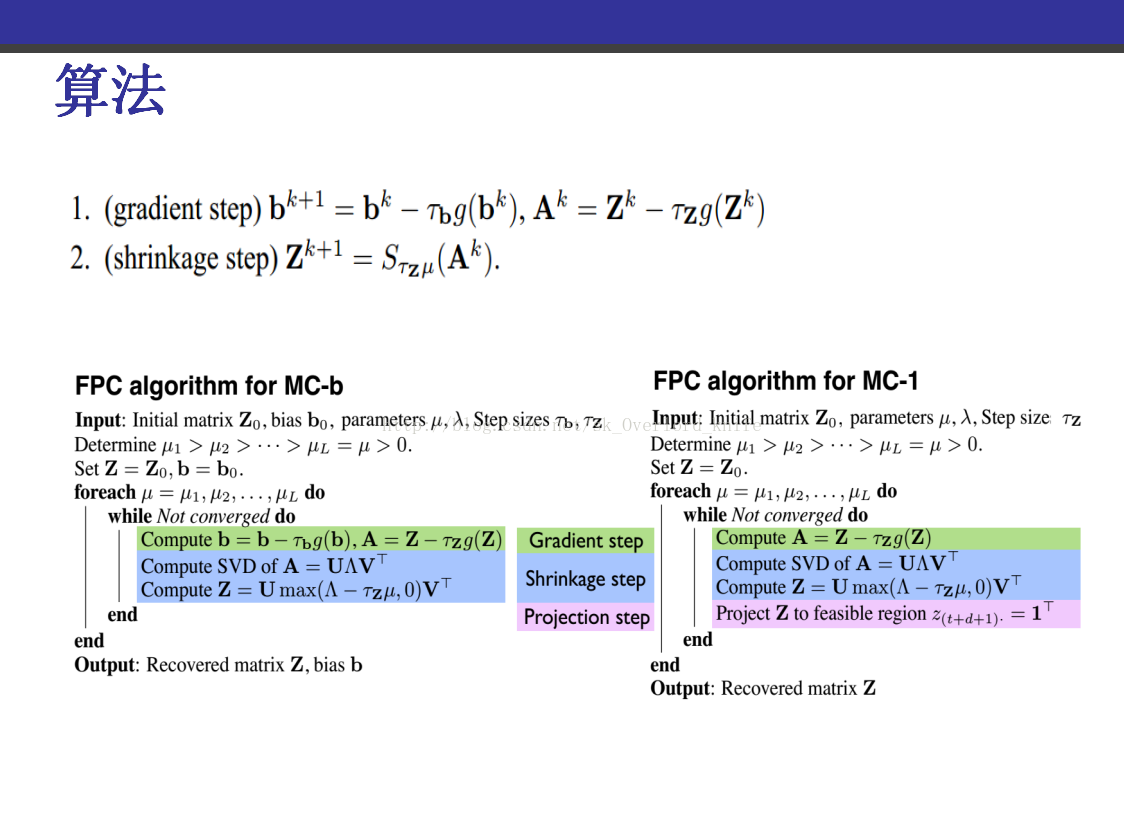
TMC.m
% - add bias term and set different lambda for output y.
%
% matrix completion FPC_kh, target output same importance: use lambda for output label and non-label
% Finds mininum mu ||X||_* + 1/2 || A(X) - b ||_2^2
% where A(X) is the projection of X onto the set of indices
% in Omega.
% For efficiency, the algorithm uses continuation (i.e. a series of
% mu, aka the "outer loop"), until mu = mu_final.
%
% Reference:
%
% "Fixed point and Bregman iterative methods for matrix
% rank minimization."
% Shiquian Ma, Donald Goldfarb, Lifeng Chen, October 2008
% ftp://ftp.math.ucla.edu/pub/camreport/cam08-78.pdf
%
% function [U,S,V,numiter] = FPC_kh3(X,Y,mu_final,lambda, maxiter,tol)
% Input: X- input features matrix (square loss function)
% Y - output label (logistic loss function) or scores matrix (square
% loss function)
% mu_final- parameter for the nuclear norm
% lambda - parameter for the output target
% maxiter controls maximum number of iterations per inner loop
% tol - target error level
% Outputs:
% U,V and Sigma are singular vectors and singular values, where
% X = U*Sigma*V'
% numiter is the number of iterations over all inner and outer loops
%
% code by Stephen Becker, srbecker@caltech.edu, March 2009
% May 2009: adding support for complex matrices
% Jan 2013: edited by Henry Thung, add parameter lambda
function [U,S,V,numiter] = TMC(n,Omega,b,mu_final,lambda, maxiter,tol,varargin)
% function [U,S,V,numiter] = FPC_kh3(X,Y,mu_final,lambda, maxiter,tol)
[lambda2, lambda3] = parseinput(varargin);
% lambda2 = 0.01;
% lambda3 = 0.01;
initial_Rank = 6;
VERBOSE = 0;
% Data assignment
OmegaX = Omega{1};
OmegaY = Omega{2};
bX = b{1};
bY = b{2};
% -- some parameters:
% GYmethod = 'logistic_mask'; % default
% GXmethod = 'regression_mask'; % default
GYmethod = 'logistic_avg'; % default
GXmethod = 'regression_avg'; % default
if strcmp(GYmethod,'logistic_mask')
tau = 2; % recommended that tau is between 1 and 2
elseif strcmp(GYmethod, 'logistic_avg');
tau = 1/min(length(OmegaX),length(OmegaY)*3.8/lambda)/2;
end
% tau = 10000;
eta_mu = 1/4; % how much to decrease mu at every step
if nargin < 7 || isempty(tol)
tol = 1e-4;
end
if nargin < 6 || isempty(maxiter)
maxiter = 500;
end
if nargin < 5 || isempty(lambda)
lambda = 2;
end
if length(n) ~= 3,
error('The length of n should be 3: Row no., X column no., Y column no!');
else
n1 = n(1); n2 = n(2); n3 = n(3); % size of matrix [X Y] where size(Y) = n1x1
end
% USE PROPACK when matrix is very big e.g. 1000x1000
% if n1*n2 < 100*100, SMALLSCALE = true; else SMALLSCALE = false; end
%
% incre = 5;
r = 1;
% s = r + 1; % estimate new rank
% normb = norm([bX; bY]);
% Initialization of X,Y,Z0
X = zeros(n1, n2);
maskX = zeros(n1, n2);
Y = zeros(n1, n3);
maskY = zeros(n1, n3);
[iX, jX] = ind2sub([n1,n2], OmegaX);
[iY, jY] = ind2sub([n1,n3], OmegaY);
for i = 1:length(OmegaX)
X(iX(i),jX(i)) = bX(i); % recovering the matrix X
maskX(iX(i),jX(i)) = 1;
end
for i = 1:length(OmegaY)
Y(iY(i),jY(i)) = bY(i); % recovering the matrix Y
maskY(iY(i),jY(i)) = 1;
end
% --- add bias term to X ----
n2 = n2+1;
X(:,n2) = ones(n1,1);
maskX(:,n2) = ones(n1,1);
% ------------------------
% spliting Y into label and regression part
Y2 = Y(:,2:end); % Y regress
maskY2 = maskY(:,2:end);
Y1 = Y(:,1); % Y label
maskY1 = maskY(:,1);
[U,Sigma,V] = svds([X Y],initial_Rank); % rank 6 initial approximation!
Z0 = U*Sigma*V';
Z = Z0;
% Gradient Calculation
ZX = Z(:,1:n2); % extract X part of the Z=[X Y]
ZY1 = Z(:,n2+1); % extract Y part of the Z=[X Y]
ZY2 = Z(:,n2+2:end); % extract Y part of the Z=[X Y]
switch GYmethod
case 'logistic_mask' % default
GY1 = -(lambda*Y1./(1+exp(Y1.*ZY1))).*maskY1; % Initial gradient of Y is same as logistic_avg
case 'logistic_avg'
GY1 = -(lambda*Y1./(1+exp(Y1.*ZY1))/length(find(Y1))).*maskY1; % gradient at Y
case 'regression_mask'
GY1 = (ZY1-Y1).*maskY1;
case 'regression_avg'
GY1 = (ZY1-Y1)/length(find(Y1)).*maskY1; % gradient at Y
otherwise
error('no such choice of GYmethod!');
end
% error test
if (length(bY)-length(find(Y1)))<0
error('Error found! inconsistent length');
end
switch GXmethod
case 'regression_mask' % initial gradient of X is same as regresion_avg
GX = lambda3*(ZX-X).*maskX; % gradient at X
GY2 = lambda2*(ZY2-Y2).*maskY2; % gradient at Y2
case 'regression_avg'
GX = lambda3*(ZX-X)/length(bX).*maskX;
GY2 = lambda2*(ZY2-Y2)/(length(bY)-length(find(Y1))).*maskY2;
otherwise
error('no such choice of GXmethod!');
end
G = [GX GY1 GY2]; % Gradient matrix
mu = normest( Z , 1e-2 );
% U = zeros(n1,1);
% V = zeros(n2+n3,1);
% S = 0;
relResid = 2;
numiter = 0;
while mu > mu_final
mu = max(mu * eta_mu,mu_final);
if VERBOSE, fprintf('FPC, mu = %f\n',mu); end
if VERBOSE == 1, fprintf('\tIteration: '); end
new_loss = 0;
% s = 2*r + 1; % estimate new rank for next iteration
for k = 1:maxiter
numiter = numiter + 1;
if VERBOSE==1, fprintf('\b\b\b\b%4d',k); end
% Gradient Calculation
% ZY = Z(:,n2); % extract Y part of the Z=[X Y]
% % GY = -(lambda*Y./(1+exp(Y.*ZY))/length(bY)).*maskY; % gradient at Y
% GY = (ZY-Y)/length(bY).*maskY; % gradient at Y
% ZX = Z(:,1:n2-1); % extract X part of the Z=[X Y]
% GX = (ZX-X)/length(bX).*maskX; % gradient at X
% G = [GX GY]; % Gradient matrix
% Perform a SVD
[U,Sigma,V] = svd(full(Z - tau*G),'econ');
% Shrink:
sigma = diag(Sigma); r = sum(sigma > tau*mu);
U = U(:,1:r); V = V(:,1:r);
sigma = sigma(1:r) - tau*mu;
S = diag(sigma);
% s = r + 1; % estimate new rank for next iteration
% update P(X) and g(X) = P*(P(X)-b)
Z = U*S*V';
Z(:,n2) = 1; % set back the value for the bias term to one.
ZX = Z(:,1:n2);
ZY1 = Z(:,n2+1); % extract Y part of the Z=[X Y]
ZY2 = Z(:,n2+2:end); % extract Y part of the Z=[X Y]
ZY = Z(:,n2+1:end);
Xc = ZX(OmegaX);
Yc = ZY(OmegaY);
nYlabel = sum(maskY(:,1)); % number of known Y label
x = [Xc;Yc(nYlabel+1:end)];
resid = x - [bX; bY(nYlabel+1:end)]; % logistic residual is not take into consideration
switch GYmethod
case 'logistic_mask' % default
GY1 = -(lambda*Y1./(1+exp(Y1.*ZY1))).*maskY1; % Initial gradient of Y is same as logistic_avg
case 'logistic_avg'
GY1 = -(lambda*Y1./(1+exp(Y1.*ZY1))/length(find(Y1))).*maskY1; % gradient at Y
case 'regression_mask'
GY1 = (ZY1-Y1).*maskY1;
case 'regression_avg'
GY1 = (ZY1-Y1)/length(find(Y1)).*maskY1; % gradient at Y
otherwise
error('no such choice of GYmethod!');
end
% error test
if (length(bY)-length(find(Y1)))<0
error('Error found! inconsistent length');
end
switch GXmethod
case 'regression_mask' % initial gradient of X is same as regresion_avg
GX = lambda3*(ZX-X).*maskX; % gradient at X
GY2 = lambda2*(ZY2-Y2).*maskY2; % gradient at Y2
case 'regression_avg'
GX = lambda3*(ZX-X)/length(bX).*maskX;
GY2 = lambda2*((ZY2-Y2)/(length(bY)-length(find(Y1)))).*maskY2;
otherwise
error('no such choice of GXmethod!');
end
% GX = (ZX-X);
G = [GX GY1 GY2]; % Gradient matrix
% G = [ZX-X ZY-Y]; % like SVT
% try
% updateSparse(G,resid,indx);
% catch
% l = lasterror;
% if strcmpi( l.identifier, 'MATLAB:UndefinedFunction')
% % mex file not installed, so do this instead:
% G = updateSparse_slow(G,resid,indx);
% else
% % some other error (unexpected)
% rethrow(lasterror)
% end
% end
% old_relResid = relResid;
% relResid = norm(resid)/normb;
normb = norm([bX; bY(nYlabel+1:end)]);
old_relResid = relResid;
relResid = norm(resid)/normb;
if VERBOSE == 2
fprintf('iteration %4d, rank is %2d, rel. residual is %.1e\n',k,r,relResid);
end
% if (relResid < tol)
% break
% end
% use stopping criteria 4.1 ("gtol")
% need to compute spectral norm
% if ~rem(k,5)
% if SMALLSCALE
% sigma_max = norm( U*V' - full(G)/mu );
% else
% A = @(x) U*(V'*x) - (G*x)/mu;
% At= @(x) V*(U'*x) - (G'*x)/mu;
% sigma_max = lansvd(A,At,n1,n2,1,'L');
% end
% if sigma_max - 1 < tol
% break
% end
% end
old_loss = new_loss;
nuclearNorm = mu*sum(diag(S));
y_loss = lambda*sum(log(1+exp(-Yc(1:nYlabel).*bY(1:nYlabel))))/nYlabel ...
+ lambda2*sum((Yc(nYlabel+1:end)-bY(nYlabel+1:end)).^2)/(2*(length(bY)-nYlabel));
x_loss = lambda3*sum((Xc-bX).^2)/(2*length(bX));
new_loss = nuclearNorm + y_loss + x_loss;
if k>1 && new_loss-old_loss > 1e-3 % break when error is increasing
break
end
% Above stopping criteria rarely applies, so add this one also:
if k > 1 && ( abs( relResid - old_relResid) / old_relResid ) < tol
break
end
if (relResid > 1e5)
disp('Divergence!');
break
end
end
if VERBOSE == 1
fprintf('\n\tRelative Residual is %.3f%%\n',100*relResid);
fprintf('\tObjective function, Y loss and X loss: %.3f, %.3f, %.3f\n',new_loss, y_loss, x_loss);
fprintf('\tRank is %d\n',r);
end
end
end
function [lambda2 lambda3] = parseinput(X)
lambda2 = 1;
lambda3 = 1;
switch length(X)
case 0 % default, do nothing
case 1
if isfield(X{1},'lambda2')
lambda2 = X{1}.lambda2;
end
if isfield(X{1},'lambda3')
lambda3 = X{1}.lambda3;
end
otherwise
if isfield(X{1},'lambda2')
lambda2 = X{1}.lambda2;
end
if isfield(X{1},'lambda3')
lambda3 = X{1}.lambda3;
end
disp('Too many input, only get lambda2 and lamnda 3, other input are ignored.')
end
end



































 940
940











 被折叠的 条评论
为什么被折叠?
被折叠的 条评论
为什么被折叠?








