数据集
https://www.physionet.org/content/challenge-2016/1.0.0/
我用的是这个数据集,如果大家以前做心音的话,也一定知道这个challenge,目前最大开源的心音数据集。我接触这个方向也是因为导师的一个项目,其实说实话就是一个简单的信号处理2分类,我自己的代码功底比较弱,比较不规范0_0。。。代码我规范之后会上传到自己的github,大家感兴趣的话可以在最下方找到。
心音处理
首先我用到的是spring的heart sound segment,直接在github上搜索就可以了,代码是matlab写的,把自己的数据集跑出来分割就行。这段代码将一个完整的心音周期分为S1,systole,S2,diastole。但是在运行代码之前要对信号下采样到1000hz。
特征提取
经常做信号分类的朋友应该知道通常我们都是把信号分成频域特征和时域特征分别进行特征提取。在心音分类任务中也是一样,我们首先对上述分成的四个段分别做时域和频域的特征提取。我在特征提取部分尝试了两种方法,一是将一段信号分割后,直接对这四个段做extraction,然后再求平均。第二种方法是把相同的段拼在一起,这样每段信号就被分成了四份,对每一份分帧之后再取平均。经过测试之后第二种的效果要明显好过第一种。特征提取之后将特征写在.csv文件里,方便查看和测试之后的分类算法。
时域:短时能量,过零率,峰度,偏振度。
频域:MFCC
http://blog.csdn.net/class_brick/article/details/82743741
不熟悉MFCC的朋友可以看看这篇文章。。。。
机器学习算法
我尝试了三种不同的分类算法:
SVM:http://blog.csdn.net/DP323/article/details/80535863
Adaboosting:https://www.cnblogs.com/fartherfuture/p/3678668.html
random forest:https://blog.csdn.net/qq_34106574/article/details/82016442
网上很多,我测试的结果如下
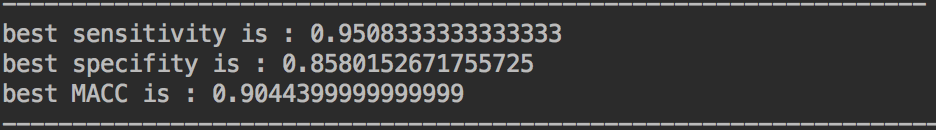
random-forest:
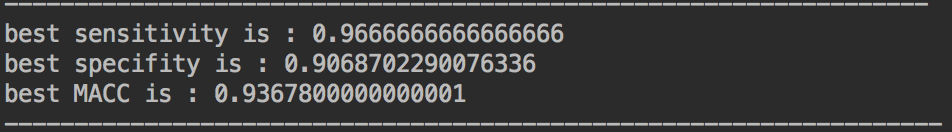
SVM:
Adaboosting:
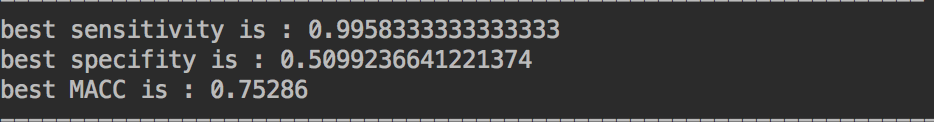
呃~Adaboosting貌似更偏向于认定一个样本是阳性,其实是件好事儿,因为现在医院不都流行保守治疗嘛。。。不过讲真这个specificity是有点太低了,后期可以再通过调整正负样本比例来进一步的优化。
随机森林+SVM
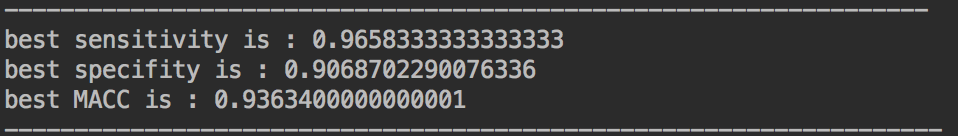
这边结合使用的ensemble learning中的stacking方法。
https://blog.csdn.net/maqunfi/article/details/82220115
网上教程很多,希望能够帮助大家理解,贴上一个。其他的模型也可以做做测试,这个大家感兴趣就自己实现吧哈哈哈。
代码:
from sklearn.ensemble import RandomForestClassifier, AdaBoostClassifier,GradientBoostingClassifier
from sklearn.model_selection import train_test_split,StratifiedKFold
from utils import caculate_MAcc
from sklearn.metrics import confusion_matrix
from sklearn.linear_model import LogisticRegression
from sklearn.model_selection import StratifiedShuffleSplit, train_test_split, StratifiedKFold, GridSearchCV
import numpy as np
from sklearn.metrics import precision_recall_fscore_support
from sklearn.metrics import roc_auc_score
from mlxtend.classifier import StackingClassifier
import os
import pandas as pd
from utils import undersampling
from sklearn import svm
from random_forest import parameter_tuning_rf,model_testing
from SVM import parameter_tuning_svm
from Adaboost import parameter_tuning_ada
from GDBT import parameter_tuning_gbdt
import joblib
import warnings
warnings.filterwarnings("ignore")
args_out = '/home/deep/heart_science/result'
epochs = 1
def pro_to(pro_arr):
not_sure = 0
pre_list = []
for i in range(len(pro_arr)):
if np.abs(pro_arr[i][0]-pro_arr[i][1]) < 0.9:
not_sure += 1
pre_list.append(0)
else:
if pro_arr[i][0] > pro_arr[i][1]:
pre_list.append(-1)
else:
pre_list.append(1)
return pre_list,not_sure
def model_training_stack(x_train, y_train, cross_val, y_name, n_maj=None, n_min=None):
sensitivity_list = []
recall_list = []
MAcc_list = []
for epoch in range(epochs):
# cross_val flag is used to specify if the model is used to test on a
# cross validation set or the blind test set
if cross_val:
# splits the training data to perform 5-fold cross validation
ss = StratifiedShuffleSplit(n_splits=5, test_size=0.2, random_state=18*epoch)
precision_list = [0]
for train_index, test_index in ss.split(x_train, y_train):
index = 0
X_train = x_train[train_index]
Y_train = y_train[train_index]
X_test = x_train[test_index]
Y_test = y_train[test_index]
#invoke the parameter tuning functiom
rf_params = parameter_tuning_rf(X_train, Y_train)
svm_params = parameter_tuning_svm(X_train, Y_train)
ada_params = parameter_tuning_ada(X_train, Y_train)
#gdbt_params = parameter_tuning_gbdt(X,Y)
rf = RandomForestClassifier(n_estimators=rf_params['n_estimators'], max_depth=rf_params['max_depth'],
n_jobs=-1,
random_state=0)
svr = svm.SVC(C=svm_params['C'], kernel='rbf', gamma=svm_params['gamma'], random_state=4, probability=True)
ada = AdaBoostClassifier(n_estimators=ada_params['n_estimators'], learning_rate=ada_params['learning_rate'],
algorithm='SAMME.R')
#gdbt = GradientBoostingClassifier(learning_rate=gdbt_params['learning_rate'],n_estimators=gdbt_params['n_estimators'],
#max_depth=gdbt_params['max_depth'],subsample=gdbt_params['subsample'],random_state = 0)
lr = LogisticRegression(C=1,max_iter=500)
clfs = [ada,rf,svr]
sclf = StackingClassifier(classifiers=clfs,use_probas=True,average_probas=False,
meta_classifier=lr)
sclf.fit(X_train,Y_train)
# intialize the random forest classifier
y_predict = sclf.predict(X_test)
#pro = sclf.predict_proba(X_test)
#y_predict,not_sure = pro_to(pro)
#precision, recall, f_score, _ = precision_recall_fscore_support(Y_test, y_predict, pos_label=1,average='binary')
c_mat = confusion_matrix(Y_test, y_predict)
se,sp,MAcc = caculate_MAcc(c_mat)
print(c_mat)
precision_list.append(se)
index += 1
sensitivity_list.append(se)
recall_list.append(sp)
MAcc_list.append(MAcc)
#uncertain_list.append(uncertain)
print('------------------------------------------------------------------')
print("best sensitivity is :",np.mean(sensitivity_list))
print("best specifity is :",np.mean(recall_list))
#print("uncertainty is :",np.mean(uncertain_list))
print("best MACC is :",np.mean(MAcc_list))
print('-------------------------------------------------------------------')
#return sclf
return np.mean(sensitivity_list),np.mean(recall_list),np.mean(MAcc_list)
if __name__ == "__main__":
#feature_select_path = '/home/deep/heart_science/teacher.txt'
model_apr = 'train-cross-val'
train_file = '/Users/mac/Desktop/heart_science/data_feature1.csv'
try:
train_feature = pd.read_csv(train_file)
except FileNotFoundError:
print("无法找到此csv文件")
train_feature.dropna(inplace=True)
feature_list = train_feature.columns.values.tolist()
feature_label = feature_list[-1]
feature_list.remove(feature_label)
'''
if os.path.exists(feature_select_path):
feature_list = []
with open(feature_select_path, 'r+') as f:
read_list = f.readlines()
for item in read_list:
feature_list.append(item.split('\n')[0])
else:
print('无法找到feature list,请重新运行特征选择')
'''
X_ = train_feature[feature_list]
Y_ = train_feature[feature_label]
n_maj = 0.47
n_min = 1.0
for i in range(8):
X, Y = undersampling(X_.values, Y_.values, majority_class=-1, minority_class=1,
maj_proportion=n_maj, min_proportion=n_min)
num_positive = 0
num_negitive = 0
for i in range(len(Y)):
if Y[i] == -1:
num_negitive += 1
else:
num_positive += 1
print("the number of negitive smaple is {0},and the number of positive sample is{1}".format(num_negitive,num_positive))
sclf = model_training_stack(X,Y,True,'feature_label', n_maj, n_min)
n_maj+=0.05
特征提取部分的代码我还需要规范一下再贴出来,这边是应用了stacking方法的分类器代码。最后一部分是当时写论文为了探究正负样本比例对各个评价指标的影响。。。。
贴一下需要用到的两个函数
import itertools
import pylab as pl
import numpy as np
from sklearn.metrics import precision_recall_curve
from sklearn.metrics import auc
def undersampling(X, Y, majority_class, minority_class, maj_proportion, min_proportion):
# fetch the row indices matching the majority and minority class
row_ind_maj = np.where(Y == majority_class)[0] #np.where()用来找出索引
row_ind_min = np.where(Y == minority_class)[0]
global maj_num
global min_num
if maj_proportion is None:
maj_num = row_ind_min.shape[0]
elif isinstance(maj_proportion,int): #int则为数量
maj_num = int(maj_proportion)
elif isinstance(maj_proportion, float): #float则为比例
maj_num = int(maj_proportion * row_ind_maj.shape[0])
if min_proportion is None:
min_num = row_ind_min.shape[0]
elif isinstance(min_proportion,int):
min_num = int(min_proportion)
elif isinstance(min_proportion, float):
min_num = int(min_proportion * row_ind_min.shape[0])
# sample the data based on the proportions of the majority and minority class
sampled_maj_indices = row_ind_maj[np.arange(0, maj_num)]
sampled_min_indices = row_ind_min[np.arange(0, min_num)]
sample_indices = np.concatenate((sampled_maj_indices, sampled_min_indices))
Y = Y[sample_indices]
X = X[sample_indices]
return X, Y
def caculate_MAcc(Matrix):
TP = Matrix[0][0]
FN = Matrix[0][1]
FP = Matrix[1][0]
TN = Matrix[1][1]
sensitivity = TP/(TP + FN)
specificity = TN/(FP + TN)
return sensitivity,specificity,np.round((sensitivity + specificity)/2,decimals=4)
到这为止就可以拿到一个performance还算不错的心音分类了,其实我在看论文的时候,其他信号的分类也可以用类似的方法来做。
贴一下我的论文:https://ieeexplore.ieee.org/abstract/document/9003008项目的github地址:https://github.com/pearchen/heart-sound
最新的代码找不到了,贴一个以前版本的可能比较乱大家凑合看吧,具体项目及实验方法也在论文中说明了,不妥善的地方还请各位给出解决方案。
小弟才疏学浅,希望能跟各位专业人士多多交流,如有不对的地方请多多指正!





















 4359
4359











 被折叠的 条评论
为什么被折叠?
被折叠的 条评论
为什么被折叠?








