Python,H5PY,大数据 (Python, H5PY, Big Data)
HDF5 is a great mechanism for storing large numerical arrays of homogenous type, for data models that can be organized hierarchically and benefit from tagging of datasets with arbitrary metadata.
HDF5是一种伟大的机制,用于存储同构类型的大型数值数组,用于可以分层组织的数据模型,并受益于使用任意元数据对数据集进行标记。
It’s quite different from SQL-style relational databases. HDF5 has quite a few organizational tricks up its sleeve, but if you find yourself needing to enforce relationships between values in various tables, or wanting to perform JOINs on your data, a relational database is probably more appropriate.
它与SQL样式的关系数据库完全不同。 HDF5具有许多组织上的技巧,但是如果您发现自己需要在各种表中的值之间建立关系,或者想要对数据执行JOIN,则关系数据库可能更合适。
HDF5 is just about perfect if you make minimal use of relational features and have a need for very high performance, partial I/O, hierarchical organization, and arbitrary metadata.
如果您很少使用关系功能,并且需要非常高性能,部分I / O,分层组织和任意元数据,那么HDF5几乎是完美的。
Hierarchical Data Format version 5, or “HDF5,” is the standard mechanism for storing large quantities of numerical data. As data volumes get larger, organization of data becomes increasingly important; features in HDF5 like-named datasets, hierarchically organized groups, and user-defined metadata “attributes” become essential to the analysis process.
分层数据格式版本5(或“ HDF5”)是用于存储大量数字数据的标准机制。 随着数据量的增加,数据的组织变得越来越重要。 HDF5命名数据集中的功能,分层组织的组以及用户定义的元数据“属性”对于分析过程至关重要。
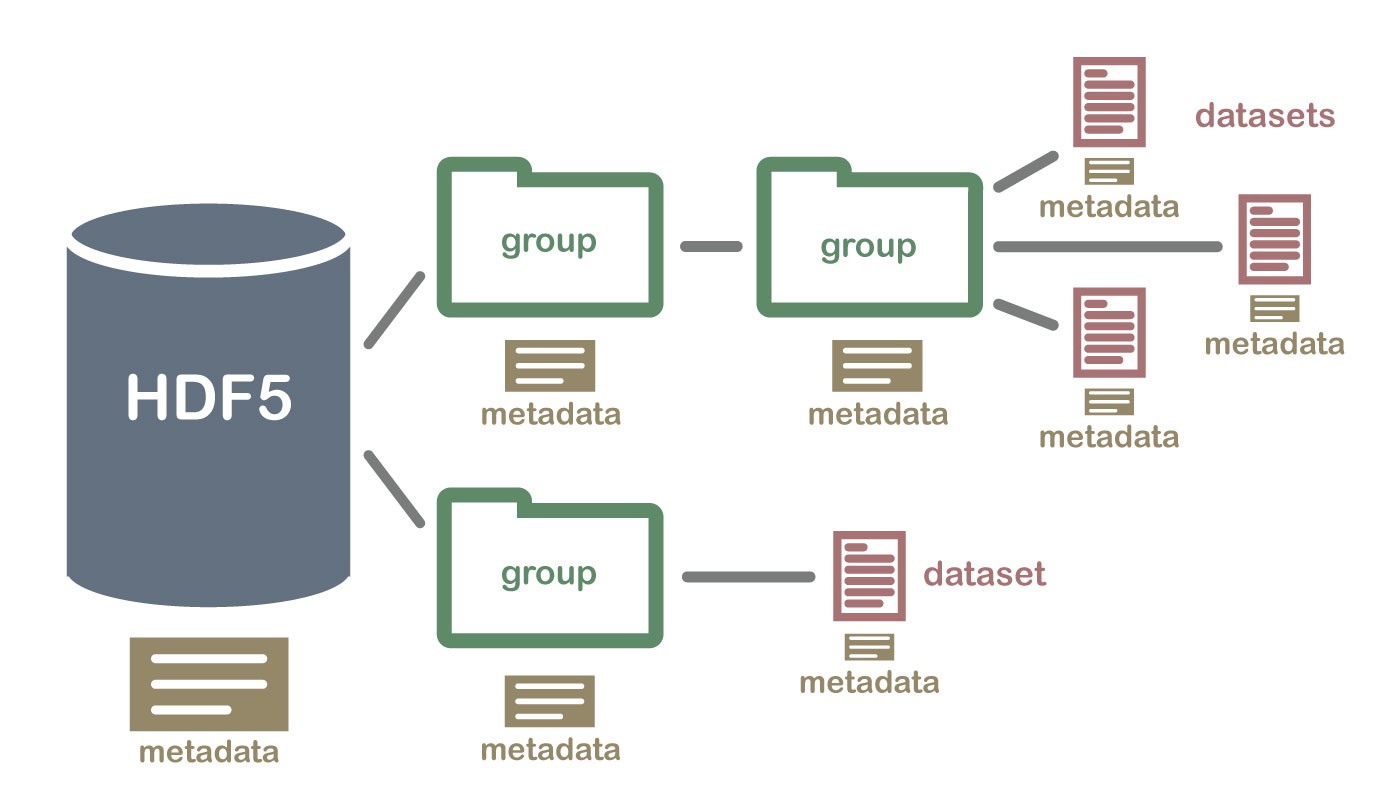
The HDF5 file is a container for two types of objects:* Datasets, which are array-like collections of data.* Groups, which are folder-like containers that hold datasets and other groups.
HDF5文件是包含两种类型对象的容器:*数据集,它们是类似于数组的数据集合。*组是类似于文件夹的包含数据集和其他组的容器。
Best of all, the files you create are in a widely-used standard binary format, which you can exchange with other people, including those who use programs like IDL and MATLAB.
最重要的是,您创建的文件采用广泛使用的标准二进制格式,您可以与其他人 (包括使用IDL和MATLAB之类的程序的人)进行交换。
H5py Python软件包 (H5py Python Package)
The h5py Python package is a Pythonic interface to the HDF5 binary data format. It helps in storing huge amounts of numerical data and easily manipulate the data from the NumPy module.
h5py Python软件包是HDF5二进制数据格式的Pythonic接口。 它有助于存储大量的数值数据,并可以轻松地从NumPy模块处理数据。
For example, We can slice into multi-terabyte datasets stored on disk, as if they were real NumPy arrays. Thousands of datasets can be stored in a single file, categorized and tagged however you want.
例如 ,我们可以将存储在磁盘上的多TB数据集切片,就像它们是真实的NumPy数组一样。 可以将成千上万个数据集存储在单个文件中,并根据需要对其进行分类和标记。
The H5py uses the NumPy and Python metaphors, like dictionary and NumPy array syntax for its operations on the data.
H5py使用NumPy和Python隐喻(例如字典和NumPy数组语法)对其数据进行操作。
For example, you can iterate over datasets in a file, or check out the .shape or .dtype attributes of datasets. You don’t need to know anything special about HDF5
例如 ,您可以遍历文件中的数据集,或检出数据集的.shape或.dtype属性。 您不需要了解有关HDF5的任何特别信息
The h5py rests on the object-oriented Cython wrapping of the HDF5 C API. Almost anything you can do from C in HDF5, you can do from h5py. The most fundamental thing to remember when using h5py is:
h5py基于HDF5 C API的面向对象Cython包装。 在HDF5中几乎可以用C进行的任何操作,都可以从h5py中进行。 使用h5py时要记住的最基本的事情是:
Groups work like dictionaries, and datasets work like NumPy arrays
组像字典一样工作,数据集像NumPy数组一样工作
安装 (Installation)
Install via pip from PyPI using the below command:
使用以下命令从PyPI通过pip安装:
pip install h5pyFor Anaconda/MiniConda, use the below command:
对于Anaconda / MiniConda,请使用以下命令:
conda install h5pyFor Canopy use the below command:
对于Canopy,请使用以下命令:
enpkg h5py打开和创建文件 (Opening & creating files)
HDF5 files also work as the standard Python file objects. They support standard modes like r/w/a, and should be closed when they are no longer in use. There is no concept of “text” vs “binary” mode.
HDF5文件也可用作标准Python文件对象。 它们支持r / w / a等标准模式,并且在不再使用时应将其关闭。 没有“文本”与“二进制”模式的概念。
import h5pyf = h5py.File('myfile.hdf5','r')组织数据和元数据 (Organizing Data and Metadata)
Below is the sample code for Weather station data that sampled the temperature, say, every 10 seconds. Using NumPy array the data is been represents for the experiment:
下面是气象站数据的示例代码,该数据每10秒采样一次温度。 使用NumPy数组可表示实验数据:
import numpy as np
temperature = np.random.random(1024)
temperatureOutputarray([ 0.44149738, 0.7407523 , 0.44243584, …, 0.19018119,
0.64844851, 0.55660748])
dt = 10.0
start_time = 1375204299 # in Unix time
station = 15
np.savez("weather.npz", data=temperature, start_time=start_time, station=station)
out = np.load("weather.npz")out["data"]Outputarray([ 0.44149738, 0.7407523 , 0.44243584, ..., 0.19018119,
0.64844851, 0.55660748])
out["start_time"]Outputarray(1375204299)out["station"]Output
array(15)So far it looks good. But what if we have more than one quantity per station? Say there’s also wind speed data to record? And suppose we have multiple stations. We could introduce some kind of naming convention.
到目前为止看起来还不错。 但是,如果我们每个站点的数量超过一个,该怎么办? 说还有风速数据要记录吗? 并假设我们有多个工作站。 我们可以引入某种命名约定。
In contrast, here’s how the application might approach storage with HDF5:
相反,这是应用程序如何使用HDF5进行存储的方式:
import h5py
f = h5py.File(“weather.hdf5”)
f[“/15/temperature”] = temperature
f[“/15/temperature”].attrs[“dt”] = 10.0
f[“/15/temperature”].attrs[“start_time”] = 1375204299
f[“/15/wind”] = wind
f[“/15/wind”].attrs[“dt”] = 5.0
----
f[“/20/temperature”] = temperature_from_station_20
----
(and so on)This example illustrates two of the important features of HDF5* organization in hierarchical groups and attributes.* Groups, like folders in a filesystem, let you store related datasets together.
此示例说明了HDF5 *组织在分层组和属性中的两个重要功能。*组,如文件系统中的文件夹,使您可以将相关的数据集存储在一起。
In this case, temperature and wind measurements from the same weather station are stored together under groups named “/15,” “/20,” etc. Attributes let you attach descriptive metadata directly to the data it describes. So if you share this file to anyone, they can easily discover the information needed to make sense of the data:
在这种情况下,来自同一气象站的温度和风的测量值将一起存储在名为“ / 15”,“ / 20”等的组下。属性使您可以将描述性元数据直接附加到其描述的数据。 因此,如果您将此文件共享给任何人,他们可以轻松地发现理解数据所需的信息:
dataset = f[“/15/temperature”]
for key, value in dataset.attrs.iteritems():
… print “%s: %s” % (key, value)
dt: 10.0
start_time: 1375204299应对大数据量 (Coping with Large Data Volumes)
As a high-level “glue” language, Python is increasingly being used for rapid visualization of big datasets and to coordinate large-scale computations that run in compiled languages like C and FORTRAN. It’s now relatively common to deal with datasets hundreds of gigabytes or even terabytes in size; HDF5 itself can scale up to exabytes.
作为一种高级“胶水”语言,Python越来越多地用于大型数据集的快速可视化以及协调以C和FORTRAN等编译语言运行的大规模计算。 现在,处理数百GB甚至是TB的数据集相对比较普遍。 HDF5本身可以扩展到EB。
On all but the biggest machines, it’s not feasible to load such datasets directly into memory. One of HDF5’s greatest strengths is its support for subsetting and partial I/O.
在除了最大的机器上的所有机器上,将此类数据集直接加载到内存中都是不可行的。 HDF5的最大优势之一就是对子集和部分I / O的支持。
For example, let’s take the 1024-element “temperature” dataset we created earlier:
例如,让我们采用我们之前创建的具有1024个元素的“温度”数据集:
dataset = f[“/15/temperature”]Here, the object named dataset is a proxy object representing an HDF5 dataset. It supports array-like slicing operations, which will be familiar to frequent NumPy users:
在此,名为数据集的对象是代表HDF5数据集的代理对象。 它支持类似数组的切片操作,这对于频繁的NumPy用户来说是很熟悉的:
dataset[0:10]Outputarray([ 0.44149738, 0.7407523 , 0.44243584, 0.3100173 , 0.04552416,
0.43933469, 0.28550775, 0.76152561, 0.79451732, 0.32603454])dataset[0:10:2]Outputarray([ 0.44149738, 0.44243584, 0.04552416, 0.28550775, 0.79451732])Keep in mind that the actual data lives on disk; when slicing is applied to an HDF5 dataset, the appropriate data is found and loaded into memory. Slicing in this fashion leverages the underlying subsetting capabilities of HDF5 and is consequently very fast.
请记住,实际数据保存在磁盘上。 将切片应用于HDF5数据集时,将找到适当的数据并将其加载到内存中。 以这种方式进行切片可利用HDF5的基本子集功能,因此速度非常快。
Another great thing about HDF5 is that you have control over how storage is allocated. For example, except for some metadata, a brand new dataset takes zero space, and by default, bytes are only used on disk to hold the data you actually write.
HDF5的另一个优点是您可以控制存储的分配方式。 例如,除了某些元数据外,全新的数据集占用的空间为零,默认情况下,字节仅在磁盘上用于保存您实际写入的数据。
For example, here’s a 2-terabyte dataset you can create on just about any computer:
例如,这是一个2 TB的数据集,您几乎可以在任何计算机上创建:
big_dataset = f.create_dataset(“big”, shape=(1024, 1024, 1024, 512),
dtype=’float32')Although no storage is yet allocated, the entire “space” of the dataset is available to us. We can write anywhere in the dataset, and only the bytes on disk necessary to hold the data are used:
尽管尚未分配存储空间,但数据集的整个“空间”可供我们使用。 我们可以在数据集中的任何地方写入数据,并且仅使用磁盘上保存数据所需的字节:
big_dataset[344, 678, 23, 36] = 42.0When storage is at a premium, you can even use transparent compression on a dataset-by-dataset basis:
当存储非常珍贵时,您甚至可以在每个数据集的基础上使用透明压缩:
compressed_dataset = f.create_dataset(“comp”, shape=(1024,), dtype=’int32', compression=’gzip’)
compressed_dataset[:] = np.arange(1024)
compressed_dataset[:]Output
array([ 0, 1, 2, …, 1021, 1022, 1023])平行HDF5 (Parallel HDF5)
Parallel HDF5 is a configuration of the HDF5 library which lets you share open files across multiple parallel processes. It uses the MPI (Message Passing Interface) standard for interprocess communication. Consequently, when using Parallel HDF5 from Python, your application will also have to use the MPI library.
并行HDF5是HDF5库的配置,可让您跨多个并行进程共享打开的文件。 它使用MPI(消息传递接口)标准进行进程间通信。 因此,当从Python使用Parallel HDF5时,您的应用程序还必须使用MPI库。
This is accomplished through the mpi4py Python package, which provides excellent, complete Python bindings for MPI.
这是通过mpi4py Python软件包完成的,该软件包为MPI提供了出色的完整Python绑定。
from mpi4py import MPIimport h5py
rank = MPI.COMM_WORLD.rank # The process ID (integer 0-3 for 4-process run)
f = h5py.File('parallel_test.hdf5', 'w', driver='mpio', comm=MPI.COMM_WORLD)
dset = f.create_dataset('test', (4,), dtype='i')
dset[rank] = rank
f.close()Run the program:
运行程序:
mpiexec -n 4 python demo2.pyLooking at the file with h5dump:
用h5dump查看文件:
h5dump parallel_test.hdf5
HDF5 "parallel_test.hdf5" {
GROUP "/" {
DATASET "test" {
DATATYPE H5T_STD_I32LE
DATASPACE SIMPLE { ( 4 ) / ( 4 ) }
DATA {
(0): 0, 1, 2, 3
}
}
}
}避免将RAM用于大型阵列-而是使用磁盘进行存储 (Avoid using RAM for Large Arrays — instead, use Disk for storage)
Avoid creating a large array saving RAM unless until required for computation else the RAM will become full. This is very much important while working on a virtual environment with limited computing environments like Kaggle Kernel, AWS, Google Colab. In such cases, it is suggested to store in disk instead of RAM.
避免创建大的阵列保存RAM,除非在计算需要之前,否则RAM将变满。 在具有有限计算环境(例如Kaggle Kernel,AWS,Google Colab)的虚拟环境中工作时,这非常重要。 在这种情况下,建议存储在磁盘中而不是RAM中。
Create a NumPy array of 600,600,600,6 and assign an index with value 100.
创建一个600,600,600,6的NumPy数组,并分配一个值为100的索引。

results = np.ones((600,600,600,6))
results[2,4,5,1] = 100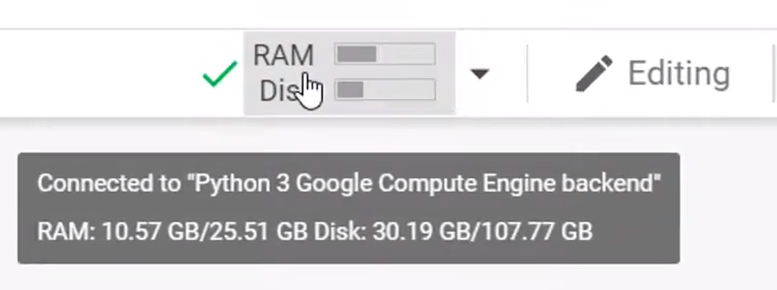
Using H5py package we can do the same thing as the above by saving the data in the disk and the operation of assigning data with RAM usage not been affected.
使用H5py包,我们可以通过将数据保存在磁盘中以及在不影响RAM使用的情况下分配数据的操作来执行与上述相同的操作。
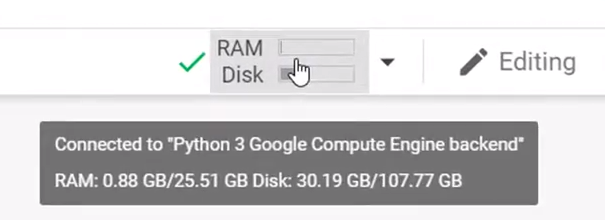
import h5py
hdf5_store = h5py.File(“./cache.hdf5”, “a”)
results = hdf5_store.create_dataset(“results”, (600,600,600,6), compression=”gzip”)
# do something…
results[2,4,5,1] = 100
We can see that the RAM usage is not been affected by using the H5py package and getting the same functionality. This way h5py helps in creating, storing, and modifying large data sets with limited computing environments.
我们可以看到,使用H5py软件包并获得相同的功能不会影响RAM的使用。 这样,h5py有助于在有限的计算环境下创建,存储和修改大型数据集。
最后的想法 (Final Thoughts)
As Python is increasingly used to handle large numerical datasets, more emphasis has been placed on the use of standard formats for data storage and communication. HDF5, the most recent version of the “Hierarchical Data Format” originally developed at the National Center for Supercomputing Applications (NCSA), has rapidly emerged as the mechanism of choice for storing scientific data in Python. At the same time, many researchers who use (or are interested in using) HDF5 have been drawn to Python for its ease of use and rapid development capabilities.
随着Python越来越多地用于处理大型数值数据集,人们越来越重视使用标准格式进行数据存储和通信。 HDF5是最初在国家超级计算应用程序中心(NCSA)上开发的“分层数据格式”的最新版本,已Swift成为选择在Python中存储科学数据的机制。 同时,许多使用(或感兴趣使用)HDF5的研究人员因其易用性和快速开发能力而被Python吸引。
其他参考 (Further References)
O’Reilly book, Python and HDF5
O'Reilly的书 , Python和HDF5





















 1070
1070











 被折叠的 条评论
为什么被折叠?
被折叠的 条评论
为什么被折叠?








