多粒度级联森林解读
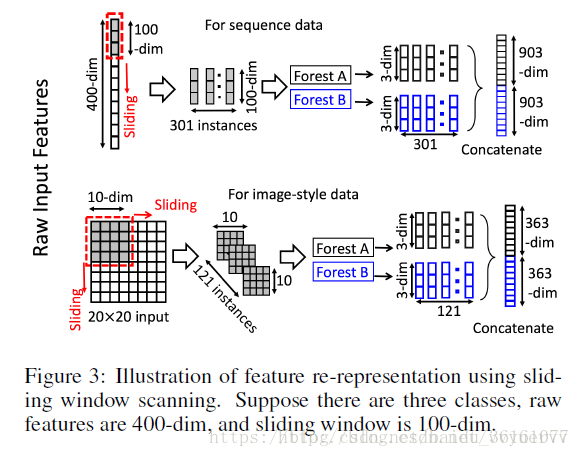
本博文简单解读一下周志华教授的多粒度级联森林算法。废话不多说,多粒度级联森林的结构主要分为两部分,一部分是多粒度扫描部分,还有就是级联森林部分。多粒度扫描结构图如下所示:
从上面的图可以看出,假设原始数据是400维的,然后分别用大小为100,200,300的滑块进行滑动,分别得到301 * 100,201 *100, 101 * 100的数据,也就是得到了301,201,101个实例为100维,200维,300维的子样本数据(instances),这些数据其实都是一个样本里面的特征,不是有301个样本,不过这里为了好理解就将这301个当成了子样本,这一步有点k折交叉采样的意思,然后分别将这批数据送到级联随机森林里面,这里的随机森林是成对出现的,一个是普通随机森林一个是完全随机森林,为了增加集成学习的多样性。完全随机森林和普通随机森林的不同点在于,完全随机森林中的每棵树通过随机选择一个特征在树的每个节点进行分割实现生成,树一直生长,直到每个叶节点只包含相同类的实例或不超过10个实例。类似地,普通每个随机森林也包含1000棵树,通过随机选择
(
d
)
\sqrt{(d)}
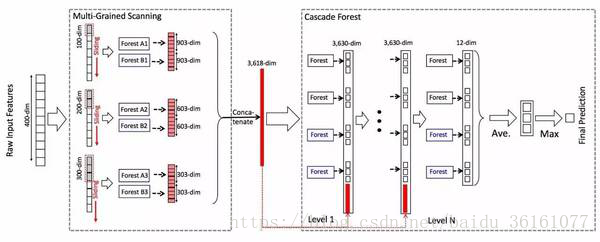
(d) 数量的特征作为候选(d是输入特征的数量),然后选择具有最佳 gini 值的特征作为分割。每个森林中的树的数值是一个超参数。假设一个随机森林的分类之后的类别个数是3个,一个样本经过一个随机森林后就得到了一个3维的概率分布,之前我们分别得到了301,201,101个样本,那么这3批样本进入一个随机森林以后得到的概率就有903,603,303个了,图中每一批次用了2个随机森林,那么最后得到的概率的个数就有了903 X 2, 603 X 2, 303 X 2.然后将这些概率拼接称一个,就变成了一个3618dim的数据了,到这里,多粒度扫描的步骤完成了,多粒度扫描的过程就相当于特征的提取,下面是级联森林的过程了
将得到的3618维的数据x送到随机森林里面,每个随机森林又得到了一个3维的数据,图中level 1用了4个随机森林,其中上面两个黑色的是完全随机森林,下面的两个是普通随机森林,这样就得到了4个3维的密度数据,再将原先的x,拼接起来就变成了3618 + 12 = 3630维的数据了,将这个数据作为下一层的输入,之后又得到4个3维的数据,再和原始的x拼接,得到3630维的术数据,作为下一层的输入,这样传递到最后一层,就不用将原来的数据再拼接起来了,因为它是最后一层了,不需要再拼接起来作为下一层的输入了,最后输出4个3维的数据,将这4个3维的数据取平均,得到一个3维的数据,然后在这个3维的数据里面取最大的,作为预测。整个多粒度级联森林的层数是自适应调节的,在级联森林的构造阶段,只要构造当前层时,经过交叉验证的验证准确率相比于前一层没有提升,那么级联森林的构造就此停止,整个结构也就完成了。
好了,基本的算法结构已经粗略的分析完了,后面还会慢慢完善的,比如随机森林的构成,决策树中的属性分裂度量方法等。上大餐了。。。。。。。。。。。。。。
#!usr/bin/env python
"""
Version : 0.1.4
Date : 15th April 2017
Author : Pierre-Yves Lablanche
Email : plablanche@aims.ac.za
Affiliation : African Institute for Mathematical Sciences - South Africa
Stellenbosch University - South Africa
License : MIT
Status : Under Development
Description :
Python3 implementation of the gcForest algorithm preesented in Zhou and Feng 2017
(paper can be found here : https://arxiv.org/abs/1702.08835 ).
It uses the typical scikit-learn syntax with a .fit() function for training
and a .predict() function for predictions.
"""
import itertools
import numpy as np
from sklearn.ensemble import RandomForestClassifier
from sklearn.model_selection import train_test_split
from sklearn.metrics import accuracy_score
__author__ = "Pierre-Yves Lablanche"
__email__ = "plablanche@aims.ac.za"
__license__ = "MIT"
__version__ = "0.1.4"
__status__ = "Development"
# noinspection PyUnboundLocalVariable
class gcForest(object):
def __init__(self, shape_1X=None, n_mgsRFtree=30, window=None, stride=1,
cascade_test_size=0.2, n_cascadeRF=2, n_cascadeRFtree=101, cascade_layer=np.inf,
min_samples_mgs=0.1, min_samples_cascade=0.05, tolerance=0.0, n_jobs=1):
'''
shape_1X:单个样本元素的形状,可以是int型的数字、元组、列表形式
n_mgsRFtree: 多粒度扫描时,随机森林中的决策树的个数,多粒度扫描阶段的决策树的个数30
window:滑动窗口
stride:滑动步长
cascade_test_size:测试集占总共几个的分数或者测试集的样本的个数
n_cascadeRF: 一个级联层中的随机森林的个数,每个完全随机森林对应了一个普通随机森林(伪随机森林)
所以一个级联层中的随机森林的个数是 2 × n_cascadeRF
n_cascadeRFtree:级联森林中单个随机森林中决策树的个数101
tolerance 如果级联森林中一级一级往下传递下去的时候,精确度的提升不再大于tolerance,那么cascade_layer就不再增长了,就停止了
n_jobs 是并行处理的随机森林的个数,在RandomForestClassifier中有n_jobs 这个参数
'''
""" gcForest Classifier.
:param shape_1X: int or tuple list or np.array (default=None)
Shape of a single sample element [n_lines, n_cols]. Required when calling mg_scanning!
For sequence data a single int can be given.
:param n_mgsRFtree: int (default=30)
Number of trees in a Random Forest during Multi Grain Scanning.
:param window: int (default=None)
List of window sizes to use during Multi Grain Scanning.
If 'None' no slicing will be done.
:param stride: int (default=1)
Step used when slicing the data.
:param cascade_test_size: float or int (default=0.2)
Split fraction or absolute number for cascade training set splitting.
:param n_cascadeRF: int (default=2)
Number of Random Forests in a cascade layer.
For each pseudo Random Forest a complete Random Forest is created, hence
the total numbe of Random Forests in a layer will be 2*n_cascadeRF.
:param n_cascadeRFtree: int (default=101)
Number of trees in a single Random Forest in a cascade layer.
:param min_samples_mgs: float or int (default=0.1)
Minimum number of samples in a node to perform a split
during the training of Multi-Grain Scanning Random Forest.
If int number_of_samples = int.
If float, min_samples represents the fraction of the initial n_samples to consider.
:param min_samples_cascade: float or int (default=0.1)
Minimum number of samples in a node to perform a split
during the training of Cascade Random Forest.
If int number_of_samples = int.
If float, min_samples represents the fraction of the initial n_samples to consider.
:param cascade_layer: int (default=np.inf)
mMximum number of cascade layers allowed.
Useful to limit the contruction of the cascade.
:param tolerance: float (default=0.0)
Accuracy tolerance for the casacade growth.
If the improvement in accuracy is not better than the tolerance the construction is
stopped.
:param n_jobs: int (default=1)
The number of jobs to run in parallel for any Random Forest fit and predict.
If -1, then the number of jobs is set to the number of cores.
"""
setattr(self, 'shape_1X', shape_1X)
setattr(self, 'n_layer', 0)
setattr(self, '_n_samples', 0)
setattr(self, 'n_cascadeRF', int(n_cascadeRF))
if isinstance(window, int):
#这里的if elif 是为了将window 处理成列表的形式
setattr(self, 'window', [window])
elif isinstance(window, list):
setattr(self, 'window', window)
setattr(self, 'stride', stride)
setattr(self, 'cascade_test_size', cascade_test_size)
setattr(self, 'n_mgsRFtree', int(n_mgsRFtree))
setattr(self, 'n_cascadeRFtree', int(n_cascadeRFtree))
setattr(self, 'cascade_layer', cascade_layer)
setattr(self, 'min_samples_mgs', min_samples_mgs)
setattr(self, 'min_samples_cascade', min_samples_cascade)
setattr(self, 'tolerance', tolerance)
setattr(self, 'n_jobs', n_jobs)
def fit(self, X, y):
""" Training the gcForest on input data X and associated target y.
:param X: np.array
Array containing the input samples.
Must be of shape [n_samples, data] where data is a 1D array.
:param y: np.array
1D array containing the target values.
Must be of shape [n_samples]
"""
if np.shape(X)[0] != len(y):
raise ValueError('Sizes of y and X do not match.')
mgs_X = self.mg_scanning(X, y)
_ = self.cascade_forest(mgs_X, y)
def predict_proba(self, X):
""" Predict the class probabilities of unknown samples X.
:param X: np.array
Array containing the input samples.
Must be of the same shape [n_samples, data] as the training inputs.
:return: np.array
1D array containing the predicted class probabilities for each input sample.
"""
mgs_X = self.mg_scanning(X)
cascade_all_pred_prob = self.cascade_forest(mgs_X)
predict_proba = np.mean(cascade_all_pred_prob, axis=0)
return predict_proba
def predict(self, X):
""" Predict the class of unknown samples X.
:param X: np.array
Array containing the input samples.
Must be of the same shape [n_samples, data] as the training inputs.
:return: np.array
1D array containing the predicted class for each input sample.
"""
pred_proba = self.predict_proba(X=X)
predictions = np.argmax(pred_proba, axis=1)
return predictions
def mg_scanning(self, X, y=None):
""" Performs a Multi Grain Scanning on input data.
:param X: np.array
Array containing the input samples.
Must be of shape [n_samples, data] where data is a 1D array.
:param y: np.array (default=None)
:return: np.array
Array of shape [n_samples, .. ] containing Multi Grain Scanning sliced data.
"""
setattr(self, '_n_samples', np.shape(X)[0]) #样本的个数
shape_1X = getattr(self, 'shape_1X') # 单个样本元素的形状
if isinstance(shape_1X, int):
# 如果shape_1X是int型的数据如,就给它加个维度
#单个样本的形状就变成了[1,shape_1X],如果shape_1X是3,那么后面就会变成[1,3]的形状,
#像[1,3]这样的数据是属于时序数据
shape_1X = [1,shape_1X]
if not getattr(self, 'window'):
setattr(self, 'window', [shape_1X[1]])
'''
这里的window是列表的形式,在论文中分别取了[100,200,300]的大小的窗口,在下面的代码中,会将
不同大小的窗口一个个取出来(int型),然后用这些大小不同的窗口去扫描原始数据。下面的win_size
就是传到方法中的参数window,上面的window 和下面的window 类型不是一样的
'''
mgs_pred_prob = [] #多粒度扫描输出的三个概率就存在这个列表里面
for wdw_size in getattr(self, 'window'): #在多粒度扫描阶段,论文的图中用了3种不同的滑动窗口
#分别是100,200,300大小的
wdw_pred_prob = self.window_slicing_pred_prob(X, wdw_size, shape_1X, y=y)#扫描
mgs_pred_prob.append(wdw_pred_prob)
return np.concatenate(mgs_pred_prob, axis=1)
def window_slicing_pred_prob(self, X, window, shape_1X, y=None):
""" Performs a window slicing of the input data and send them through Random Forests.
If target values 'y' are provided sliced data are then used to train the Random Forests.
:param X: np.array
Array containing the input samples.
Must be of shape [n_samples, data] where data is a 1D array.
:param window: int
Size of the window to use for slicing.
:param shape_1X: list or np.array
Shape of a single sample.
:param y: np.array (default=None)
Target values. If 'None' no training is done.
:return: np.array
Array of size [n_samples, ..] containing the Random Forest.
prediction probability for each input sample.
"""
n_tree = getattr(self, 'n_mgsRFtree')#每个随机森林中决策树的个数
min_samples = getattr(self, 'min_samples_mgs')#多粒度扫描时最小的样本数或者最小占比0.1
stride = getattr(self, 'stride')# 步长
if shape_1X[0] > 1:# 如果样本数组的行数大于1,可以判断为是图像数据
print('Slicing Images...')
sliced_X, sliced_y = self._window_slicing_img(X, window, shape_1X, y=y, stride=stride)# 图像扫描
else:#否则就是时序数据
print('Slicing Sequence...')
sliced_X, sliced_y = self._window_slicing_sequence(X, window, shape_1X, y=y, stride=stride)#时序数据扫描
if y is not None:#当样本有标签时
n_jobs = getattr(self, 'n_jobs')# 0
#扫描完成后进入随机森林,经过随机森林后得到3维的数据
prf = RandomForestClassifier(n_estimators=n_tree, max_features='sqrt',
min_samples_split=min_samples, oob_score=True, n_jobs=n_jobs)
# 搭建随机森林框架
crf = RandomForestClassifier(n_estimators=n_tree, max_features=None,
min_samples_split=min_samples, oob_score=True, n_jobs=n_jobs)
print('Training MGS Random Forests...')
prf.fit(sliced_X, sliced_y)# 将扫描得到的数据送到随机森林里面,随机森林有两种
crf.fit(sliced_X, sliced_y)# crf是普通随机森林 common
setattr(self, '_mgsprf_{}'.format(window), prf)
setattr(self, '_mgscrf_{}'.format(window), crf)
pred_prob_prf = prf.oob_decision_function_
pred_prob_crf = crf.oob_decision_function_
if hasattr(self, '_mgsprf_{}'.format(window)) and y is None:
prf = getattr(self, '_mgsprf_{}'.format(window))
crf = getattr(self, '_mgscrf_{}'.format(window))
pred_prob_prf = prf.predict_proba(sliced_X)
pred_prob_crf = crf.predict_proba(sliced_X)
pred_prob = np.c_[pred_prob_prf, pred_prob_crf]
return pred_prob.reshape([getattr(self, '_n_samples'), -1])
def _window_slicing_img(self, X, window, shape_1X, y=None, stride=1):
""" Slicing procedure for images
:param X: np.array
Array containing the input samples.
Must be of shape [n_samples, data] where data is a 1D array.
:param window: int
Size of the window to use for slicing.
:param shape_1X: list or np.array
Shape of a single sample [n_lines, n_cols].
:param y: np.array (default=None)
Target values.
:param stride: int (default=1)
Step used when slicing the data.
:return: np.array and np.array
Arrays containing the sliced images and target values (empty if 'y' is None).
"""
if any(s < window for s in shape_1X):
#图像数据的两个维度都小于窗口的大小,那就报错
raise ValueError('window must be smaller than both dimensions for an image')
len_iter_x = np.floor_divide((shape_1X[1] - window), stride) + 1#属性方向滑块滑动之后得到的维度
#假设图片为20X20,滑块大小为10x10,步长为1,滑动之后得到的维度就是len = (20 - 10) / 1 + 1 = 11
len_iter_y = np.floor_divide((shape_1X[0] - window), stride) + 1
iterx_array = np.arange(0, stride*len_iter_x, stride)
itery_array = np.arange(0, stride*len_iter_y, stride)
ref_row = np.arange(0, window)#这里传进来的window是一个int型数据
ref_ind = np.ravel([ref_row + shape_1X[1] * i for i in range(window)])
inds_to_take = [ref_ind + ix + shape_1X[1] * iy
for ix, iy in itertools.product(iterx_array, itery_array)]
#index to take 取的像素点索引
'''
如果图像的像素点的位置可以用数字来表示的话.比如一张图片的像素是 15X15,用5X5的滑动块
来取图片的像素,那么得到的索引就是[[array([0,1,2,3,4,15,16,17,18,19,30,31,32,
33,34,45,46,47,48,49,60,61,62,63,64,65]),array(......)]],这组数字就代表了
需要被卷积出来的像素点的位置,一行有15个,第一行是0 -- 14,最后一行是第15行,最后一个
像素点的索引号是15 ** 2 - 1 = 244
'''
sliced_imgs = np.take(X, inds_to_take, axis=1).reshape(-1, window**2)
#列向取像素点,将取到的像素点展平,再重新弄成窗口的大小: window ** 2
#shape_1X是列表,只包含两个元素[len(row),len(column)],X是原始图片
sliced_target = np.repeat(y, len_iter_x * len_iter_y)
'''
这里的y是label,上面我们用类似卷积的方式从图像中获得了特征信息,横向走了len_iter_x次,
列向走了len_iter_y次,这样我们就总共的到了36个像素块,每个像素块的大小都是window * window的
我们将每块像素块都赋予标签信息,为了后面随机森林的拟合,repeat是复制
'''
return sliced_imgs, sliced_target
def _window_slicing_sequence(self, X, window, shape_1X, y=None, stride=1):
""" Slicing procedure for sequences (aka shape_1X = [.., 1]).
:param X: np.array
Array containing the input samples.
Must be of shape [n_samples, data] where data is a 1D array.
:param window: int
Size of the window to use for slicing.
:param shape_1X: list or np.array
Shape of a single sample [n_lines, n_col].
:param y: np.array (default=None)
Target values.
:param stride: int (default=1)
Step used when slicing the data.
:return: np.array and np.array
Arrays containing the sliced sequences and target values (empty if 'y' is None).
"""
if shape_1X[1] < window:
raise ValueError('window must be smaller than the sequence dimension')
len_iter = np.floor_divide((shape_1X[1] - window), stride) + 1
iter_array = np.arange(0, stride*len_iter, stride)
ind_1X = np.arange(np.prod(shape_1X))
inds_to_take = [ind_1X[i:i+window] for i in iter_array]
sliced_sqce = np.take(X, inds_to_take, axis=1).reshape(-1, window)
sliced_target = np.repeat(y, len_iter)
return sliced_sqce, sliced_target
def cascade_forest(self, X, y=None):# 级联森林阶段
""" Perform (or train if 'y' is not None) a cascade forest estimator.
:param X: np.array
Array containing the input samples.
Must be of shape [n_samples, data] where data is a 1D array.
:param y: np.array (default=None)
Target values. If 'None' perform training.
:return: np.array
1D array containing the predicted class for each input sample.
"""
if y is not None:# if y is not None,表示的就是进入训练阶段,测试阶段送到forest是不需要送label的
setattr(self, 'n_layer', 0)
test_size = getattr(self, 'cascade_test_size')# 0.2 测试集占数据集的比例
max_layers = getattr(self, 'cascade_layer') # cascade_layer = np.inf
#这里用了最大的网络深度是无限大,就是让网络自己调整深度,等到精度增长不再变化了,因为此代码中的tolerance = 0
tol = getattr(self, 'tolerance')
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=test_size)
self.n_layer += 1
prf_crf_pred_ref = self._cascade_layer(X_train, y_train)
accuracy_ref = self._cascade_evaluation(X_test, y_test)
feat_arr = self._create_feat_arr(X_train, prf_crf_pred_ref)#传进来的原始数据集和经过随机森林后得到的概率向量拼接
self.n_layer += 1
prf_crf_pred_layer = self._cascade_layer(feat_arr, y_train)
accuracy_layer = self._cascade_evaluation(X_test, y_test)#准确度提升
while accuracy_layer > (accuracy_ref + tol) and self.n_layer <= max_layers:
accuracy_ref = accuracy_layer
prf_crf_pred_ref = prf_crf_pred_layer
feat_arr = self._create_feat_arr(X_train, prf_crf_pred_ref)
self.n_layer += 1
prf_crf_pred_layer = self._cascade_layer(feat_arr, y_train)
accuracy_layer = self._cascade_evaluation(X_test, y_test)
elif y is None:
at_layer = 1
prf_crf_pred_ref = self._cascade_layer(X, layer=at_layer)
while at_layer < getattr(self, 'n_layer'):
at_layer += 1
feat_arr = self._create_feat_arr(X, prf_crf_pred_ref)
prf_crf_pred_ref = self._cascade_layer(feat_arr, layer=at_layer)
return prf_crf_pred_ref
def _cascade_layer(self, X, y=None, layer=0):
""" Cascade layer containing Random Forest estimators.
If y is not None the layer is trained.
:param X: np.array
Array containing the input samples.
Must be of shape [n_samples, data] where data is a 1D array.
:param y: np.array (default=None)
Target values. If 'None' perform training.
:param layer: int (default=0)
Layer indice. Used to call the previously trained layer.
:return: list
List containing the prediction probabilities for all samples.
"""
n_tree = getattr(self, 'n_cascadeRFtree')# 101 级联森林中每个随机森林中决策树的个数
n_cascadeRF = getattr(self, 'n_cascadeRF') # 2
min_samples = getattr(self, 'min_samples_cascade') # 0
n_jobs = getattr(self, 'n_jobs') # 0
prf = RandomForestClassifier(n_estimators=n_tree, max_features='sqrt',
min_samples_split=min_samples, oob_score=True, n_jobs=n_jobs) # 普通随机森林
crf = RandomForestClassifier(n_estimators=n_tree, max_features=None,
min_samples_split=min_samples, oob_score=True, n_jobs=n_jobs)
# 上面的prf 和 crf只是搭建好了随机森林的框架,并没有送入数据
prf_crf_pred = []
if y is not None:# 有标签label
print('Adding/Training Layer, n_layer={}'.format(self.n_layer))
for irf in range(n_cascadeRF):
prf.fit(X, y)
crf.fit(X, y)
setattr(self, '_casprf{}_{}'.format(self.n_layer, irf), prf)
#指明prf,crf是级联森林中的那一层的随机森林的prf和crf
setattr(self, '_cascrf{}_{}'.format(self.n_layer, irf), crf)
prf_crf_pred.append(prf.oob_decision_function_)
prf_crf_pred.append(crf.oob_decision_function_)
elif y is None:# 无标签
for irf in range(n_cascadeRF):
prf = getattr(self, '_casprf{}_{}'.format(layer, irf))
crf = getattr(self, '_cascrf{}_{}'.format(layer, irf))
prf_crf_pred.append(prf.predict_proba(X))
prf_crf_pred.append(crf.predict_proba(X))
return prf_crf_pred#得到的是经过每个随机森林后得到的一个3维概率向量的拼接
def _cascade_evaluation(self, X_test, y_test): # 计算准确率
""" Evaluate the accuracy of the cascade using X and y.
:param X_test: np.array
Array containing the test input samples.
Must be of the same shape as training data.
:param y_test: np.array
Test target values.
:return: float
the cascade accuracy.
"""
casc_pred_prob = np.mean(self.cascade_forest(X_test), axis=0)
casc_pred = np.argmax(casc_pred_prob, axis=1)
casc_accuracy = accuracy_score(y_true=y_test, y_pred=casc_pred)
print('Layer validation accuracy = {}'.format(casc_accuracy))
return casc_accuracy
def _create_feat_arr(self, X, prf_crf_pred):
""" Concatenate the original feature vector with the predicition probabilities
of a cascade layer.
向量拼接,将原始的x和经过随机森立后得到的向量进行拼接
:param X: np.array
Array containing the input samples.
Must be of shape [n_samples, data] where data is a 1D array.
:param prf_crf_pred: list
Prediction probabilities by a cascade layer for X.
:return: np.array
Concatenation of X and the predicted probabilities.
To be used for the next layer in a cascade forest.
"""
swap_pred = np.swapaxes(prf_crf_pred, 0, 1)
add_feat = swap_pred.reshape([np.shape(X)[0], -1])
feat_arr = np.concatenate([add_feat, X], axis=1)
return feat_arr
经过本人的检验,这个算法的代码是极好内存的,因为在多粒度扫描部分,通过多粒度扫描,相当于充分提取了样本的特征,但是同时带来的是子样本的增加同时也会带来计算内存的猛涨。目前该代码还不是适应于维度太大地数据或者样本量太大的数据,虽然本算法本来就是针对小样本学习的。而且这个代码只能适用于单个通道的图像数据,不适用于多个通道的图像数据。另外提一下,编写这个代码的P.Y人很好,有问必答。大家可以主动提问。
上面这些图是代码中存在的一些缺陷,读者可以根据需要进行相应的研究和调整。























 4629
4629











 被折叠的 条评论
为什么被折叠?
被折叠的 条评论
为什么被折叠?








