首先,感谢Chai_zheng博主提供的代码思路,本文代码基于该博主思路所编写!!!感谢这个开源的时代。
一、贝叶斯原理:
作为一名研究(虽然是菜鸟级别的,哈哈),那还是用学习贝叶斯的人员来自称吧,还是要吹一下贝叶斯的几大优点的。
贝叶斯网络是一种概率图形模型,广泛应用于各种人工智能推理和因果建模中。在众多的分类模型中,贝叶斯网络对处理不确定性推理有着明显的优势。它主要用概率刻画因果关系,并可以处理存在噪声信息或者数据缺失的数据集。最重要的是贝叶斯网络是一种简单的、可解释分类模型,可在多场合提供决策支持。贝叶斯网络分为结构学习和参数学习两个部分。结构学习是形成一个有向无环图(DAG)的过程,图中节点代表属性,有向边代表属性间的条件关系或因果关系。参数学习以条件概率分布刻画属性对其父节点的依赖关系,通过样本的频率表示概率并填充条件概率表。从1988年Pearl提出贝叶斯网络后,经过近三十年的发展形成了很多新算法,具有代表性的有朴素贝叶斯(Naive Bayes,NB)、树增广朴素贝叶斯模型(Tree-Augmented Naive Bayes,TAN)和K阶贝叶斯分类模型(k-dependence Bayesian Cla-ssifier,KDB)【后面的算法打算自己慢慢的用python实现,敬请期待】
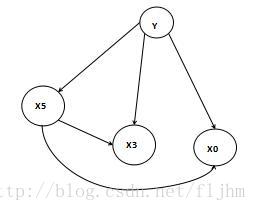
进入正题,在贝叶斯理论中有一个十分重要的假设就是条件独立性假设,该假设可以描述为除了父节点外,节点Xi与其他节点无依赖关系(即相互独立),这一点是贝叶斯研究成立的树根,为了便于理解画个草图(如图1)。X5只与Y有关,X3只与X5和Y有关,与X3相互独立,但对于朴素贝叶斯来说非类节点之间相互独立(即没有弧)。
图1
贝叶斯网络能处理离散数据和连续数据,不同的平台处理这两种数据利用的原理大致相同,利用条件独立性假设,离散数据的条件概率计算公式如下:
P(Xi|Y)=P(Y)∏P(Xi|Y)
对于任意给定的随机样本集x =(x1 ,x2 ,··· ,xn ),其对应的联合概率分布表示如下:
P(c,x)
= P(c)P(x1|c)P(x2 |c)···P(xn |c)
= P(c)∏P(x i |c) (i=1...N)
其中P(c)为类先验概率.
对于连续数据用概率密度函数来计算P(Xi|C),其中μ,σ分别代表数据的均值和标准差
二、建模的基本思路
1、计算先验概率,离散的就计算条件概率,连续的就计算概率密度
2、根据训练集训练模型(朴素贝叶斯就是得到先验概率Pc,均值,标准差)
3、根据测试集测试模型,和真实标签值进行比较,预测对了就累加matchCount
4、matchCount/测试集总数据= 准确率
三,python代码如下
(代码最后自己敲一下没这样才有意义,有些地方不懂的自己打印出来看看,理解了才是重点)
好看的代码千篇一律,各自的理解万里无一。(源自:好看皮囊千篇一律,有趣的灵魂万里无一,^_^)
采用葡萄酒(wine.txt)数据集进行测试,取测试集占比0.4,在72个测试样本上正确率为95%-100%(抽取样本的随机性导致结果不同),足见其实用性的是下载地址链接: https://pan.baidu.com/s/1i4E78VF 密码: 5u8x 别人可能需要积分,这个不需要积分,百度分享,本就是uci上捐赠的数据集,【如有侵权请及时私信】
# coding=utf-8 import math import time import numpy as np from collections import Counter from sklearn import preprocessing from sklearn.model_selection import train_test_split def calcProbDensity(meanLabel, stdLabel, test_X): numAttributes = len(test_X) MultiProbDensity = 1.0 print "this is calcPD" # print meanLabel # print stdLabel for i in range(numAttributes): MultiProbDensity *= np.exp(-np.square(test_X[i] - meanLabel[i]) / (2.0 * np.square(stdLabel[i]))) / ( np.sqrt(2.0 * math.pi) * stdLabel[i]) print MultiProbDensity return MultiProbDensity def calcPriorProb(Y_train): # 计算先验概率P(c) i, j = 0, 0 global labelValue, classNum numSamples = Y_train.shape[0] # 读取Y_train的第一维度的长度,比如多少行。 labelValue = np.zeros((numSamples, 1)) # 用前i行来保存标签值 # print "laV:",labelValue Y_train_counter = sum(Y_train.tolist(), []) # 将Y_train转化为可哈希的数据结构 cnt = Counter(Y_train_counter) # 计算标签值的类别个数{1,2,3}及各类样例的个数 # print "cnt",cnt for key in cnt: labelValue[i] = key i += 1 classNum = i # print "laV2:",labelValue # print classNum Pc = np.zeros((classNum, 1)) # 不同类的先验概率 eachLabelNum = np.zeros((classNum, 1)) # 每类样例数,多少行多少列,这里是classNum行1列 for key in cnt: Pc[j] = float(cnt[key]) / numSamples #这里加float是为了避免/整除号使Pc=0 eachLabelNum[j] = cnt[key] j += 1 return labelValue, eachLabelNum, classNum, Pc def trainBayes(X_train, Y_train): startTime = time.time() numTrainSamples, numAttributes = X_train.shape print "trainBayes" # print numAttributes,numTrainSamples labelValue, eachLabelNum, classNum, Pc = calcPriorProb(Y_train) meanlabelX, stdlabelX = [], [] # 存放每一类样本在所有属性上取值的均值和方差 for i in range(classNum): k = 0 labelXMaxtrix = np.zeros((int(eachLabelNum[i]), numAttributes)) for j in range(numTrainSamples): if Y_train[j] == labelValue[i]: labelXMaxtrix[k] = X_train[j, :] k += 1 meanlabelX.append(np.mean(labelXMaxtrix, axis=0).tolist()) # 求该矩阵的列均值与无偏标准差,append至所有类 stdlabelX.append(np.std(labelXMaxtrix, ddof=1, axis=0).tolist()) meanlabelX = np.array(meanlabelX).reshape(classNum, numAttributes) stdlabelX = np.array(stdlabelX).reshape(classNum, numAttributes) # print meanlabelX # print "###" # print stdlabelX print('---Train completed.Took %f s.' % ((time.time() - startTime))) return meanlabelX, stdlabelX, Pc def predict(X_test, Y_test, meanlabelX, stdlabelX, Pc): numTestSamples = X_test.shape[0] matchCount = 0 for m in range(X_test.shape[0]): x_test = X_test[m, :] # 轮流取测试样本 pred = np.zeros((classNum, 1)) # 对不同类的概率 print "Pc:", Pc for i in range(classNum): pred[i] = calcProbDensity(meanlabelX[i, :], stdlabelX[i, :], x_test) * Pc[i] # 计算属于各类的概率 # print i print pred predict1 = labelValue[np.argmax(pred)] # 取最大的类标签,np.argmax(pred)返回最大数据所在的index print predict1, "####", Y_test[m] if predict1 == Y_test[m]: matchCount += 1 print "matchCount", matchCount, numTestSamples accuracy = float(matchCount) / numTestSamples #这里加float是为了避免/整除号使accuracy=0 # print accuracy return accuracy if __name__ == '__main__': print('Step 1.Loading data...') # 数据集下载http://download.csdn.net/download/chai_zheng/10009919 data = np.loadtxt("data/wine.txt", delimiter=',') # 载入葡萄酒数据集 print('---Loading completed.') x = data[:, 1:14] y = data[:, 0].reshape(178, 1) # print x # print "!!!!" # print y print('Step 2.Splitting and preprocessing data...') X_train, X_test, Y_train, Y_test = train_test_split(x, y, test_size=0.4) # 拆分数据集 scaler = preprocessing.StandardScaler().fit(X_train) # 数据标准化 X_train = scaler.transform(X_train) X_test = scaler.transform(X_test) print('---Splittinging completed.\n---Number of training samples:%d\n---Number of testing samples:%d' \ % (X_train.shape[0], X_test.shape[0])) print('Step 3.Training...') meanlabelX, stdlabelX, Pc = trainBayes(X_train, Y_train) print('Step 4.Testing...') accuracy = predict(X_test, Y_test, meanlabelX, stdlabelX, Pc) print('---Testing completed.Accuracy:%.3f%%' % (accuracy * 100))
结果截图:











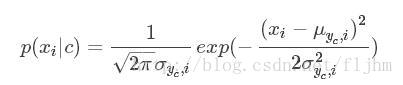















 845
845

 被折叠的 条评论
为什么被折叠?
被折叠的 条评论
为什么被折叠?








