自从4月份成功部署模型,实现自动分割算法落地应用后,便马不停蹄地致力于改进网络,提高分割精度,期间学习并复现了几种网络与实现了三维体数据的DataLoader和Cropping,记录一下以便不时之需;
BraTS2018-No.1
网络结构
首先BraTS2018第一名方案的backbone与CNN类似,不同的是加入了decoder和VAE路径,前者结合Unet的skip concat作解码,还原到与原图像size相等的patch后,在softmax层上完成多分类任务,大体上与Unet相差无几;于是可以照葫芦画瓢地写出编码路径和解码路径的structure。
编码路径-卷积块
class EncoderBlock(nn.Module):
def __init__(self, inChans, outChans, stride=1, padding=1, num_groups=8, activation="relu", normalizaiton="group_normalization"):
super(EncoderBlock, self).__init__()
if normalizaiton == "group_normalization":
self.norm1 = nn.GroupNorm(num_groups=num_groups, num_channels=inChans)
self.norm2 = nn.GroupNorm(num_groups=num_groups, num_channels=inChans)
if activation == "relu":
self.actv1 = nn.ReLU(inplace=True)
self.actv2 = nn.ReLU(inplace=True)
elif activation == "elu":
self.actv1 = nn.ELU(inplace=True)
self.actv2 = nn.ELU(inplace=True)
self.conv1 = nn.Conv3d(in_channels=inChans, out_channels=outChans, kernel_size=3, stride=stride, padding=padding)
self.conv2 = nn.Conv3d(in_channels=inChans, out_channels=outChans, kernel_size=3, stride=stride, padding=padding)
def forward(self, x):
residual = x
out = self.norm1(x)
out = self.actv1(out)
out = self.conv1(out)
out = self.norm2(out)
out = self.actv2(out)
out = self.conv2(out)
out += residual
return out
值得注意的是卷积块是用了Residual Block,我们只需将x与out相加即可。
解码路径–卷积块
大体上与编码路径的block没有差别,唯一需要注意的是GN操作与Conv3d操作中,需要将输入通道数对于改成输出通道数(也可以用反卷积来完成,说不定也是个tricks)
self.norm1 = nn.GroupNorm(num_groups=num_groups, num_channels=outChans)
self.norm2 = nn.GroupNorm(num_groups=num_groups, num_channels=outChans)
self.conv1 = nn.Conv3d(in_channels=inChans, out_channels=outChans, kernel_size=3, stride=stride, padding=padding)
self.conv2 = nn.Conv3d(in_channels=outChans, out_channels=outChans, kernel_size=3, stride=stride, padding=padding)
下采样
偷懒一点可以使用maxpooling,借助torch自带的函数很容易实现,可能需要调整参数,我这里是kernel=3, stride=2, padding=1
也可以自行实现downsample,这里使用了卷积操作完成,目的是为了提取更多的low level information,同时防止过拟合,使用了dropout层。
class DownSampling(nn.Module):
def __init__(self, inChans, outChans, stride=2, kernel_size=3, padding=1, dropout_rate=None):
super(DownSampling, self).__init__()
self.dropout_flag = False
self.conv1 = nn.Conv3d(in_channels=inChans,
out_channels=outChans,
kernel_size=kernel_size,
stride=stride,
padding=padding,
bias=False)
if dropout_rate is not None:
self.dropout_flag = True
self.dropout = nn.Dropout3d(dropout_rate,inplace=True)
def forward(self, x):
out = self.conv1(x)
if self.dropout_flag:
out = self.dropout(out)
return out
Trick: VAE实现
这部分一开始没想好怎么去实现论文里的正态分布,查了一圈,发现torch.distributions已经贴心的为你写好了前向传播和后向求导过程:
def VDraw(x):
return torch.distributions.Normal(x[:,:128], x[:,128:]).sample()
接下来是对VAE的采样过程进行重现:
class VDResampling(nn.Module):
def __init__(self, inChans=256, outChans=256, dense_features=(10,12,8), stride=2, kernel_size=3, padding=1, activation="relu", normalizaiton="group_normalization"):
super(VDResampling, self).__init__()
midChans = int(inChans / 2)
self.dense_features = dense_features
if normalizaiton == "group_normalization":
self.gn1 = nn.GroupNorm(num_groups=8,num_channels=inChans)
if activation == "relu":
self.actv1 = nn.ReLU(inplace=True)
self.actv2 = nn.ReLU(inplace=True)
elif activation == "elu":
self.actv1 = nn.ELU(inplace=True)
self.actv2 = nn.ELU(inplace=True)
self.conv1 = nn.Conv3d(in_channels=inChans, out_channels=16, kernel_size=kernel_size, stride=stride, padding=padding)
self.dense1 = nn.Linear(in_features=16*dense_features[0]*dense_features[1]*dense_features[2], out_features=inChans)
self.dense2 = nn.Linear(in_features=midChans, out_features=midChans*dense_features[0]*dense_features[1]*dense_features[2])
self.up0 = LinearUpSampling(midChans,outChans)
def num_flat_features(self, x):
size = x.size()[1:]
num_features = 1
for s in size:
num_features *= s
return num_features
def forward(self, x):
out = self.gn1(x)
out = self.actv1(out)
out = self.conv1(out)
out = out.view(-1, self.num_flat_features(out))#为重构张量的维度,相当于numpy中resize的功能
out_vd = self.dense1(out)
distr = out_vd
out = VDraw(out_vd) #正态分布处理
out = self.dense2(out)
out = self.actv2(out)
out = out.view((1, 128, self.dense_features[0],self.dense_features[1],self.dense_features[2]))
out = self.up0(out)
return out, distr
剩下的decoder与上面所述的差距无几,可能有点出入的是结构图的上采样过程不能直接用解码block,这里提供用于前向传播时,一种线性上采样的思路:
nn.functional.interpolate(out, scale_factor=2, mode=trilinear, align_corners=True)
训练结果图:

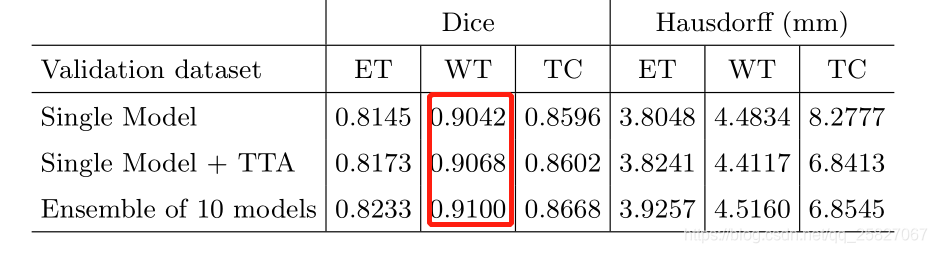
目前只训练了40个epoch,validation loss 与论文描述还是有点点差距,可能接着训练就能达到预期的效果了,比3D UNET好用,第一名确实名不虚传;(就是很费时间训练)
dataset->h5
以往的博客里,我一般是2D图像一张张读入,然后相应地,从原数据集和label中去除全0的图片,最后做完预处理后交给模型进行训练,但这样带来的问题也是显而易见的:
1、285分数据全部读入后,算按8:2比例分割成训练集和验证集,都还是将近有150k张240*240的图片,并且GPU的RAM不允许batch_size过大,一般最多是8(加上模型满打满算刚好20GB);
2、脑肿瘤分割中的一大难点:正负样本不平衡,模型在学习的过程中,计算一张张梯度更新权值时产生的计算开销、FLOPS也是很大的,况且就算做出来取得不错的结果,在z方向上,没有考虑上下切片的空间位置关系,精度也会有一定的差距。
当然2D网络也是有优点的,就是能快速验证一个模块能否提高精度。
扯远了,本章节是借鉴Isensee在2017年提供的开源代码上进行改动,实现三维读入。
def fetch_training_data_files(return_subject_ids=False):
training_data_files = list()
subject_ids = list()
processed_dir = config["img_dir"]
for idx, subject_dir in enumerate(glob.glob(os.path.join(processed_dir, "*"))):
subject_ids.append(os.path.basename(subject_dir))
subject_files = list()
for modality in config["training_modalities"] + ["seg"]:
subject_files.append(os.path.join(subject_dir, os.path.basename(subject_dir) + '_' + modality + ".nii"))
training_data_files.append(tuple(subject_files))
if return_subject_ids:
return training_data_files, subject_ids
else:
return training_data_files
以上代码段是从硬盘中读取文件名,检索成list,目的是为了将其统一读入,转换成h5文件类型,其中对于不同的数据集路径或者文件命名不同的情况,对这句代码进行调整即可:
subject_files.append(os.path.join(subject_dir, os.path.basename(subject_dir) + '_' + modality + ".nii"))
























 1422
1422

 被折叠的 条评论
为什么被折叠?
被折叠的 条评论
为什么被折叠?








