0、论文背景
本论文主要是通过改进的BPSO算法来进行特征选择,特征选择的效果通过1-NN分类算法来检验。通过比较得出提出的算法在特征选择上具有一定的优越性。
Zhang Y, Gong D, Hu Y, et al. Feature selection algorithm based on bare bones particle swarm optimization[J]. Neurocomputing, 2015, 148: 150-157.
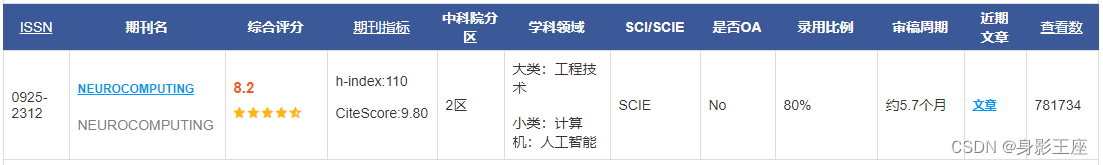
是一篇二区的论文,个人觉得针对特征选择的实际问题而提出的改进的BPSO算法实际上创新性不是太大,但是可以让我们就相关问题有一个清晰的认识。
1、论文的核心思想
本文提出了一种利用BPSO求解最优特征子集的新方法,称为 binary BPSO。该算法(binary BPSO)设计了(a reinforced memory strategy)一种增强记忆策略来更新粒子的局部最优值,以避免粒子中突出基因的退化,并提出了一种统一的组合算法( uniform combination (UC))来平衡局部开发和全局探索。此外,利用1-NN方法作为分类器来评价粒子的分类精度。本文选取了一些国际标准数据集,对该算法进行了评价。
有关Binary PSO算法请参见:









 订阅专栏 解锁全文
订阅专栏 解锁全文
















 613
613











 被折叠的 条评论
为什么被折叠?
被折叠的 条评论
为什么被折叠?










