图分类,图机器学习最新进展
1.Flat_Pooling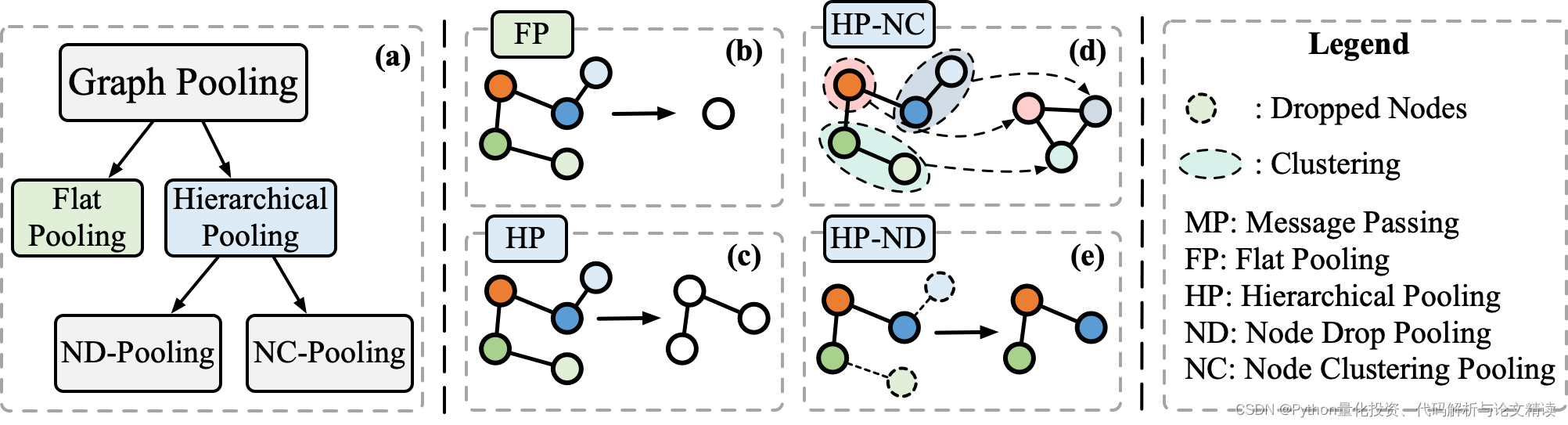
| Title | Venue | Task | Code | Dataset |
|---|---|---|---|---|
| DMLAP: Multi-level attention pooling for graph neural networks: Unifying graph representations with multiple localities | Neural Networks 2022 | 1. Graph Classification | None | synthetic, OGB-molhiv, OGB-ppa, MCF-7 (TU dataset) |
| GraphTrans: Representing Long-Range Context for Graph Neural Networks with Global Attention 🌟 | NIPS 2021 | 1. Graph Classification | 1.PyTorch | NCI1, NCI109, code2, molpcba |
| GMT: Accurate Learning of Graph Representations with Graph Multiset Pooling. 🌟 | ICLR 2021 | 1. Graph Classification 2. Graph Reconstruction 3. Graph Generation | 1.PyTorch 2.PyTorch-Geometric | D&D, PROTEINS, MUTAG, IMDB-B, IMDB-M, COLLAB, OGB-MOLHIV, OGB-Tox21, OGB-ToxCast, OGB-BBBP, ZINC(Reconstruction), QM9(Generation) |
| QSGCNN: Learning Graph Convolutional Networks based on Quantum Vertex Information Propagation | TKDE 2021 | 1. Graph Classification | None | MUTAG, PTC, NCI1, PROTEINS, D&D, COLLAB, IMDB-B, IMDB-M, RED-B |
| DropGNN: DropGNN: Random Dropouts Increase the Expressiveness of Graph Neural Networks | NIPS 2021 | 1. Graph Classification 2. Graph Regression | PyTorch | MUTAG, PTC, PROTEINS, IMDB-B, IMDB-M QM9(Regression) |
| SSRead: Learnable Structural Semantic Readout for Graph Classification | ICDM 2021 | 1. Graph Classification | PyTorch | D&D, MUTAG, Mutagencity, NCI1,PROTEINS, IMDB-B, IMDB-M |
| FlowPool: Pooling Graph Representations with Wasserstein Gradient Flows | ArXiv 2021 | 1. Graph Classification | None | BZR, COX2, PROTEINS |
| DKEPool: Distribution Knowledge Embedding for Graph Pooling | TKDE 2022 | 1. Graph Classification | PyTorch | IMDB-B, IMDB-M, MUTAG, PTC, NCI1, PROTEINS, REDDIT-BINARY, OGB-MOLHIV, OGB-BBB |
| FusionPooling: Hybrid Low-order and Higher-order Graph Convolutional Networks | Computational Intelligence and Neuroscience 2020 | 1. Text Classification 2. node classification | None | 20-Newsgroups // Cora, CiteSeer, PubMed |
| SOPool: Second-Order Pooling for Graph Neural Networks | TPAMI 2020 | 1. Graph Classification | None | MUTAG, PTC PROTEINS, NCI1, COLLAB, IMDB-B, IMDB-M, REDDIT-BINARY,REDDIT-MULTI |
| StructSa: Structured self-attention architecture for graph-level representation learning | Pattern Recognition 2020 | 1. Graph Classification | None | MUTAG, PTC PROTEINS, NCI1, COLLAB, IMDB-B, IMDB-M, REDDIT-BINARY,REDDIT-MULTI |
| NAS: Graph Neural Network Architecture Search for Molecular Property Prediction | ICBD 2020 | 1. Graph Regression | None | QM7, QM8, QM9, ESOL, FreeSolv, Lipophilicity |
| Neural Pooling for Graph Neural Networks | ArXiv 2020 | 1. Graph Classification | None | MUTAG, PTC PROTEINS, NCI1, COLLAB, IMDB-B, IMDB-M, REDDIT-BINARY,REDDIT-MULTI-5K |
| GFN: Are Powerful Graph Neural Nets Necessary? A Dissection on Graph Classification | ArXiv 2019 | 1. Graph Classification | PyTorch | MUTAG, PROTEINS, D&D, NCI1, ENZYMES, IMDB-B, IMDB-M, RDT-B. REDDTIT-Multi-5K, REDDIT-Multi-12K, COLLAB |
| GIN: How Powerful are Graph Neural Networks? | ICLR 2019 | 1. Graph Classification | PyTorch | MUTAG, PROTEINS, PTC, NCI1, IMDB-B, IMDB-M, RDT-B. RDT-Multi-5K, COLLAB |
| Semi-Supervised Graph Classification: A Hierarchical Graph Perspective | WWW 2019 | 1. Graph Classification | PyTorch | Tencent |
| DEMO-Net: Degree-specific Graph Neural Networks for Node and Graph Classification | KDD 2019 | 1. Graph Classification | TensorFlow | MUTAG, PTC PROTEINS,ENZYMES |
| MSNAPool: Unsupervised Inductive Graph-Level Representation Learning via Graph-Graph Proximity | IJCAI 2019 | 1. Graph Classification 2. Graph similarity ranking 3. Graph visualization | TensorFlow | PTC, IMDB-B, WEB, NCI109, REDDIT-Multi-12K |
| PiNet: Attention Pooling for Graph Classification | NIPS-W 2019 | 1. Graph Classification | Code | MUTAG, PTC, NCI1, NCI109, PROTEINS, Erdõs-Rényi graphs |
| DAGCN: Dual Attention Graph Convolutional Networks | IJCNN 2019 | 1. Graph Classification | PyTorch | NCI1, D&D, ENZYMES, NCI109, PROTEINS, PTC |
| DeepSet: Universal Readout for Graph Convolutional Neural Networks | IJCNN 2019 | 1. Graph Classification | Code | MUTAG, PTC, NCI1, PROTEINS,D&D |
| SortPool: An End-to-End Deep Learning Architecture for Graph Classification | AAAI 2018 | 1. Graph Classification | 1.PyTorch-Geometric, 2.Matlab, 3.PyTorch 4.Spektral | MUTAG, PTC, NCI1 PROTEINS, D&D |
| Set2set: Order Matters: Sequence to Sequence for Sets | ICLR 2016 | - | PyTorch-Geometric | - |
| GatedPool: Gated Graph Sequence Neural Networks | ICLR 2016 | - | PyTorch-Geometric | - |
| DCNN: Diffusion-Convolutional Neural Networks | NIPS 2016 | 1. Graph Classification | Theano | NCI1, NCI109, MUTAG, PCI, ENZYMES |
Hierarchical_Pooling - Node_Clustering_Pooling
| Title | Venue | Task | Code | Dataset |
|---|---|---|---|---|
| Maximal Independent Vertex Set Applied to Graph Pooling | CIKM 2022 | 1. Graph Classification | None | PROTEINS, NCI1, D&D, ENZYMES |
| Higher-order Clustering and Pooling for Graph Neural Networks | CIKM 2022 | 1. Graph Classification 2. Node Clustering | 1.PyTorch | PROTEINS, NCI1, D&D, MUTAGEN., Reddit-B, Cox2-MD, ER-MD, b-hard // Cora, PubMed, DBLP, Coauthor CS ,Amazon Photo, Amazon PC, Polblogs, Eu-email |
| Unsupervised Hierarchical Graph Pooling via Substructure-Sensitive Mutual Information Maximization | CIKM 2022 | 1. Graph Classification | None | MUTAG, PROTEINS, PTC, HIV, IMDB-B, IMDB-M |
| Structural Entropy Guided Graph Hierarchical Pooling | ICML 2022 | 1. Graph Classification 2. Node Classification 3. Graph Reconstruction | 1. PyTorch | MUTAG, PROTEINS, D&D, PTC, NCI1,IMDB-B, IMDB-M // Cora, Citeseer, Pubmed // synthetic datasets (grid and circle) |
| HGCN:Unsupervised Learning of Graph Hierarchical Abstractions with Differentiable Coarsening and Optimal Transport | AAAI 2021 | 1. Graph Classification | 1. PyTorch | MUTAG, PROTEINS, D&D, NCI109,IMDB-B, IMDB-M |
| Hierarchical Graph Representation Learning with Local Capsule Pooling | MMAsia 2021 | 1. Graph Classification 2. Graph Reconstruction | 1. PyTorch | MUTAG, PROTEINS, D&D, PTC, NCI1,IMDB-B, IMDB-M //synthetic datasets (grid and circle) |
| HGCN:Hierarchical Graph Capsule Network | AAAI 2021 | 1. Graph Classification | 1. PyTorch | MUTAG, NCI1, PROTEINS, D&D, ENZYMES, PTC, NCI109,IMDB-B, IMDB-M, Reddit-BINARY |
| HAP: Hierarchical Adaptive Pooling by Capturing High-order Dependency for Graph Representation Learning | TKDE 2021 | 1. Graph Classification 2. Graph Matching 3. Graph Similarity Learning | None | IMDB-B, IMDB-M, COLLAB, MUTAG, PROTEINS, PTC // synthetic datasets (graph matching) // AIDS, LINUX (graph similarity) |
| LCP: Hierarchical Graph Representation Learning with Local Capsule Pooling | MMAsia | 1. Graph Classification 2. Graph Reconstruction | None | D&D, PROTEINS, IMDB-B, IMDB-M, NCI1, NIC109 |
| MxPool: Multiplex Pooling for Hierarchical Graph Representation Learning | ArXiv 2021 | 1.Graph Classification | None | D&D, ENZYMES, PROTEINS, NCI109, COLLAB, RDT-MULTI |
| HIBPool: Structure-Aware Hierarchical Graph Pooling using Information Bottleneck | IJCNN 2021 | 1. Graph Classification | 1.PyTorch | ENZYMES, DD, PROTEINS, NCI1, NCI109,FRANKENSTEIN |
| MLC-GCN: Graph convolutional networks with multi-level coarsening for graph classification | Knowledge-Based Systems 2020 | 1.Graph Classification | None | D&D, ENZYMES, MUTAG, PROTEINS,IMDB-BINARY, IMDB-MULTI, REDDIT- BINARY, REDDIT-MULTI-5K |
| DGM: Deep Graph Mapper: Seeing Graphs through the Neural Lens | NIPS-W 2020 | 1. Graph Classification 2. Graph Visualisation | 1.PyTorch | D&D, PROTEINS, COLLAB, REDDIT-B |
| MuchGNN: Multi-Channel Graph Neural Networks | IJCAI 2020 | 1. Graph Classification | None | PTC, DD, PROTEINS, COLLAB, IMDB-BINARY, IMDB-MULTI, REDDIT-MULTI-12K |
| MinCutPool: Spectral Clustering with Graph Neural Networks for Graph Pooling | ICML 2020 | 1. Graph Classification 2. Graph Regression | 1.PyTorch-Geometric, 2.PyTorch | D&D, PROTEINS, COLLAB, REDDIT-BINARY, Mutagenicity, QM9(regression) |
| HaarPool: Haar Graph Pooling | ICML 2020 | 1. Graph Classification 2. Graph Regression | 1.PyTorch | MUTAG, PROTEINS, NCI1, NCI109, MUTAGEN, TRIANGLES, QM7(regression) |
| MemPool: Memory-Based Graph Networks | ICLR 2020 | 1. Graph Classification 2. Graph Regression | 1.PyTorch-Geometric, 2.PyTorch | D&D, PROTEINS, COLLAB, REDDIT-BINARY,ENZYMES ESOL(reg), Lipophilicity(reg) |
| StructPool: Structured Graph Pooling via Conditional Random Fields | ICLR 2020 | 1.Graph Classification | 1. PyTorch | ENZYMES, PTC, MUTAG, PROTEINS, COLLAB, IMDB-B, IMDB-M |
| MathNet: Haar-Like Wavelet Multiresolution-Analysis for Graph Representation and Learning | ArXiv 2020 | 1.Graph Classification 2. Graph Regression | None | D&D, PROTEINS, MUTAG, ENZYMES // QM7 (regression) MUTA-GENICITY |
| ProxPool: Graph Pooling with Node Proximity for Hierarchical Representation Learning | ArXiv 2020 | 1.Graph Classification | None | D&D, PROTEINS, NCI1, NCI109, MUTA-GENICITY |
| CliquePool: Clique pooling for graph classification | ICLR-W 2019 | 1. Graph Classification | None | D&D PROTEINS, ENZYMES, COLLAB |
| NMF: A Non-Negative Factorization approach to node pooling in Graph Convolutional Neural Networks | AIIA 2019 | 1. Graph Classification | None | D&D, PROTEINS, NCI1, ENZYMES, COLLAB |
| GRAHIES: Multi-Scale Graph Representation Learning with Latent Hierarchical Structure | CogMI 2019 | 1. Node Classification | None | Cora, CiteSeer, PubMed |
| EigenPool: Graph Convolutional Networks with EigenPooling | KDD 2019 | 1. Graph Classification | 1.PyTorch | D&D, PROTEINS, NCI1, NCI109, MUTAG, |
参考链接:https://github.com/LiuChuang0059/graph-pooling-papers#flat_pooling
https://ieeexplore.ieee.org/stamp/stamp.jsp?tp=&arnumber=9460814
























 被折叠的 条评论
为什么被折叠?
被折叠的 条评论
为什么被折叠?










