UnicodeEncodeError Traceback (most recent call last)
Cell In[3], line 146
137 if name in param_grids:
138 grid_search = GridSearchCV(
139 estimator=model,
140 param_grid=param_grids[name],
(...)
144 verbose=1
145 )
--> 146 grid_search.fit(X_train, y_train)
148 best_models[name] = grid_search.best_estimator_
149 print(f"Best params: {grid_search.best_params_}")
File ~\AppData\Roaming\Python\Python312\site-packages\sklearn\base.py:1365, in _fit_context.<locals>.decorator.<locals>.wrapper(estimator, *args, **kwargs)
1358 estimator._validate_params()
1360 with config_context(
1361 skip_parameter_validation=(
1362 prefer_skip_nested_validation or global_skip_validation
1363 )
1364 ):
-> 1365 return fit_method(estimator, *args, **kwargs)
File ~\AppData\Roaming\Python\Python312\site-packages\sklearn\model_selection\_search.py:979, in BaseSearchCV.fit(self, X, y, **params)
967 fit_and_score_kwargs = dict(
968 scorer=scorers,
969 fit_params=routed_params.estimator.fit,
(...)
976 verbose=self.verbose,
977 )
978 results = {}
--> 979 with parallel:
980 all_candidate_params = []
981 all_out = []
File D:\anacondaxiaz\Lib\site-packages\joblib\parallel.py:1347, in Parallel.__enter__(self)
1345 self._managed_backend = True
1346 self._calling = False
-> 1347 self._initialize_backend()
1348 return self
File D:\anacondaxiaz\Lib\site-packages\joblib\parallel.py:1359, in Parallel._initialize_backend(self)
1357 """Build a process or thread pool and return the number of workers"""
1358 try:
-> 1359 n_jobs = self._backend.configure(n_jobs=self.n_jobs, parallel=self,
1360 **self._backend_args)
1361 if self.timeout is not None and not self._backend.supports_timeout:
1362 warnings.warn(
1363 'The backend class {!r} does not support timeout. '
1364 "You have set 'timeout={}' in Parallel but "
1365 "the 'timeout' parameter will not be used.".format(
1366 self._backend.__class__.__name__,
1367 self.timeout))
File D:\anacondaxiaz\Lib\site-packages\joblib\_parallel_backends.py:538, in LokyBackend.configure(self, n_jobs, parallel, prefer, require, idle_worker_timeout, **memmappingexecutor_args)
534 if n_jobs == 1:
535 raise FallbackToBackend(
536 SequentialBackend(nesting_level=self.nesting_level))
--> 538 self._workers = get_memmapping_executor(
539 n_jobs, timeout=idle_worker_timeout,
540 env=self._prepare_worker_env(n_jobs=n_jobs),
541 context_id=parallel._id, **memmappingexecutor_args)
542 self.parallel = parallel
543 return n_jobs
File D:\anacondaxiaz\Lib\site-packages\joblib\executor.py:20, in get_memmapping_executor(n_jobs, **kwargs)
19 def get_memmapping_executor(n_jobs, **kwargs):
---> 20 return MemmappingExecutor.get_memmapping_executor(n_jobs, **kwargs)
File D:\anacondaxiaz\Lib\site-packages\joblib\executor.py:42, in MemmappingExecutor.get_memmapping_executor(cls, n_jobs, timeout, initializer, initargs, env, temp_folder, context_id, **backend_args)
39 reuse = _executor_args is None or _executor_args == executor_args
40 _executor_args = executor_args
---> 42 manager = TemporaryResourcesManager(temp_folder)
44 # reducers access the temporary folder in which to store temporary
45 # pickles through a call to manager.resolve_temp_folder_name. resolving
46 # the folder name dynamically is useful to use different folders across
47 # calls of a same reusable executor
48 job_reducers, result_reducers = get_memmapping_reducers(
49 unlink_on_gc_collect=True,
50 temp_folder_resolver=manager.resolve_temp_folder_name,
51 **backend_args)
File D:\anacondaxiaz\Lib\site-packages\joblib\_memmapping_reducer.py:540, in TemporaryResourcesManager.__init__(self, temp_folder_root, context_id)
534 if context_id is None:
535 # It would be safer to not assign a default context id (less silent
536 # bugs), but doing this while maintaining backward compatibility
537 # with the previous, context-unaware version get_memmaping_executor
538 # exposes too many low-level details.
539 context_id = uuid4().hex
--> 540 self.set_current_context(context_id)
File D:\anacondaxiaz\Lib\site-packages\joblib\_memmapping_reducer.py:544, in TemporaryResourcesManager.set_current_context(self, context_id)
542 def set_current_context(self, context_id):
543 self._current_context_id = context_id
--> 544 self.register_new_context(context_id)
File D:\anacondaxiaz\Lib\site-packages\joblib\_memmapping_reducer.py:569, in TemporaryResourcesManager.register_new_context(self, context_id)
562 new_folder_name = (
563 "joblib_memmapping_folder_{}_{}_{}".format(
564 os.getpid(), self._id, context_id)
565 )
566 new_folder_path, _ = _get_temp_dir(
567 new_folder_name, self._temp_folder_root
568 )
--> 569 self.register_folder_finalizer(new_folder_path, context_id)
570 self._cached_temp_folders[context_id] = new_folder_path
File D:\anacondaxiaz\Lib\site-packages\joblib\_memmapping_reducer.py:585, in TemporaryResourcesManager.register_folder_finalizer(self, pool_subfolder, context_id)
578 def register_folder_finalizer(self, pool_subfolder, context_id):
579 # Register the garbage collector at program exit in case caller forgets
580 # to call terminate explicitly: note we do not pass any reference to
581 # ensure that this callback won't prevent garbage collection of
582 # parallel instance and related file handler resources such as POSIX
583 # semaphores and pipes
584 pool_module_name = whichmodule(delete_folder, 'delete_folder')
--> 585 resource_tracker.register(pool_subfolder, "folder")
587 def _cleanup():
588 # In some cases the Python runtime seems to set delete_folder to
589 # None just before exiting when accessing the delete_folder
(...)
594 # because joblib should only use relative imports to allow
595 # easy vendoring.
596 delete_folder = __import__(
597 pool_module_name, fromlist=['delete_folder']
598 ).delete_folder
File D:\anacondaxiaz\Lib\site-packages\joblib\externals\loky\backend\resource_tracker.py:179, in ResourceTracker.register(self, name, rtype)
177 """Register a named resource, and increment its refcount."""
178 self.ensure_running()
--> 179 self._send("REGISTER", name, rtype)
File D:\anacondaxiaz\Lib\site-packages\joblib\externals\loky\backend\resource_tracker.py:196, in ResourceTracker._send(self, cmd, name, rtype)
192 if len(name) > 512:
193 # posix guarantees that writes to a pipe of less than PIPE_BUF
194 # bytes are atomic, and that PIPE_BUF >= 512
195 raise ValueError("name too long")
--> 196 msg = f"{cmd}:{name}:{rtype}\n".encode("ascii")
197 nbytes = os.write(self._fd, msg)
198 assert nbytes == len(msg)
UnicodeEncodeError: 'ascii' codec can't encode characters in position 18-19: ordinal not in range(128)
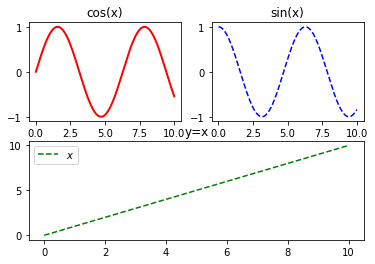





 这篇博客介绍了如何使用matplotlib的subplot()函数创建多个子图,并在每个子图中分别绘制正弦、余弦和线性函数。通过subplot(221)、subplot(222)和subplot(212)创建了三个子图,展示了不同函数的图形,并利用plot()函数进行绘制。最后,保存图像为'image.png',分辨率为100dpi,并显示图像。
这篇博客介绍了如何使用matplotlib的subplot()函数创建多个子图,并在每个子图中分别绘制正弦、余弦和线性函数。通过subplot(221)、subplot(222)和subplot(212)创建了三个子图,展示了不同函数的图形,并利用plot()函数进行绘制。最后,保存图像为'image.png',分辨率为100dpi,并显示图像。


















 1278
1278

 被折叠的 条评论
为什么被折叠?
被折叠的 条评论
为什么被折叠?










