ALEC是老师和师兄师姐发布的一篇关于主动学习的源代码,论文地址。
主动学习:
是介于监督学习和非监督学习之间的一种特殊情况。由于标记样本需要耗费大量的人力和时间,所以提出了主动学习,获取部分样本的标签,期望能得到不弱于监督学习的效果。
与半监督学习的联系与区别:
联系:主动学习和半监督学习都是从未标记的样本实例中挑选部分价值量高的样例标注后补充到已标记样例集中来提高分类器精准度,降低标注样本的工作量。
区别:主动学习需要人工进行精准标注,不会引入错误类标;半监督学习是通过具有一定分类精度的基准分类器实现对未标注样例自动标注,降低了标注代价,但不能保证标注结果完全正确。
ALEC算法学习过程:
Step 1. 在数据集中用密度(重要性)和距离获取代表性(代表性=密度*距离),并进行降序排序;
Step 2. 设当前数据集中有
N
N
N个实例对象,当前数据集可查询
N
\sqrt N
N个对象的标签;
Step 3. 如果查询的
N
\sqrt N
N个对象的标签都相同,则认为当前的簇为纯的,并将剩余对象设为同一类;
Step 4. 需要将当前数据集划分成两个子集,分别对两个子集执行 Step 3.
算法理解:
重要性(密度):围着你转的人越多,就说明你就越重要,同理,如果某个对象周围的实例越多,说明它的重要性就越高,例如一个重要性很高的实例的标签为1,那它周围实例的标签大概率也为1;
距离:是你到你领导之间的距离,如果你离他越远,他能管你的地方就越少,就是“天高皇帝远”,同理,一个实例离它的master越远,就说明它的独立性就越高,越能凸显它自己的特点;
代表性(=重要性*距离):代表性越大的实例越能代表该实例的周围结点和同一个master的其它实例。
先把老师的完整代码附上:
package activelearning;
import java.io.FileReader;
import java.util.Arrays;
import weka.core.Instances;
public class Alec {
/**
* The whole data set.
*/
Instances dataset;
/**
* The maximal number of queries that can be provided.
*/
int maxNumQuery;
/**
* The actual number of queries.
*/
int numQuery;
/**
* The radius,which is employed for density computation.
*/
double radius;
/**
* The densities of instances.
*/
double[] densities;
/**
* distance to master.
*/
double[] distanceToMaster;
/**
* Sorted indices, where the first element indicates the instance with the
* biggest density.
*/
int[] descendantDensities;
/**
* Priority.
*/
double[] priority;
/**
* The maximal distance between any pair of points.
*/
double maximalDistance;
/**
* Record the master.
*/
int[] masters;
/**
* Predicted labels
*/
int[] predictedLabels;
/**
* Instance status. 0 for unprocessed, 1 for queried, 2 for classified.
*/
int[] instanceStatusArray;
/**
* The descendant indices to show the representativeness of instances in a
* descendant order.
*/
int[] descendantRepresentatives;
/**
* Indicate the cluster of each instance. It is only used in
* clusterInTwo(int[]);
*/
int[] clusterIndices;
/**
* Blocks with size no more than this threshold should not be split further.
*/
int smallBlockThreshold = 3;
/**
*********************
* The constructor.
*
* @param paraFilename The filename.
*********************
*/
public Alec(String paraFilename) {
try {
FileReader tempFileReader = new FileReader(paraFilename);
dataset = new Instances(tempFileReader);
dataset.setClassIndex(dataset.numAttributes() - 1);
tempFileReader.close();
} catch (Exception ee) {
System.out.println(ee);
System.exit(0);
} // Of try
computeMaximalDistance();
clusterIndices = new int[dataset.numInstances()];
}// Of the constructor.
/**
********************
* Compute the maximal distance. The result is stored in a member variable.
*********************
*/
public void computeMaximalDistance() {
maximalDistance = 0;
double tempDistance;
for (int i = 0; i < dataset.numInstances(); i++) {
for (int j = 0; j < dataset.numInstances(); j++) {
tempDistance = distance(i, j);
if (maximalDistance < tempDistance) {
maximalDistance = tempDistance;
} // Of if
} // Of for j
} // Of for i
System.out.println("maximalDistance = " + maximalDistance);
}// Of computeMaximalDistance
/**
*********************
* The Euclidean distance between two instances. Other distance measures
* unsupported for simplicity.
*
*
* @param paraI The index of the first instance.
* @param paraJ The index of the second instance.
* @return The distance.
*********************
*/
public double distance(int paraI, int paraJ) {
double resultDistance = 0;
double tempDifference;
for (int i = 0; i < dataset.numAttributes() - 1; i++) {
tempDifference = dataset.instance(paraI).value(i) - dataset.instance(paraJ).value(i);
resultDistance += tempDifference * tempDifference;
} // Of for i
resultDistance = Math.sqrt(resultDistance);
return resultDistance;
}// Of distance
/**
**********************************
* Cluster based active learning.
*
* @param paraRatio The ratio of the maximal distance.
* @param paraMaxNumQuery The maximal number of queries for the whole
* data set.
* @param paraSmallBlockThreshold The small block threshold.
**********************************
*/
public void clusterBasedActiveLearning(double paraRatio, int paraMaxNumQuery, int paraSmallBlockThreshold) {
radius = maximalDistance * paraRatio;
smallBlockThreshold = paraSmallBlockThreshold;
maxNumQuery = paraMaxNumQuery;
predictedLabels = new int[dataset.numInstances()];
for (int i = 0; i < dataset.numInstances(); i++) {
predictedLabels[i] = -1;
} // Of for i
computeDensitiesGaussian();
computeDistanceToMaster();
computePriority();
descendantRepresentatives = mergeSortToIndices(priority);
System.out.println("descendantRepresentatives = " + Arrays.toString(descendantRepresentatives));
numQuery = 0;
clusterBasedActiveLearning(descendantRepresentatives);
}// Of clusterBasedActiveLearning
/**
********************
* Compute the densities using Gaussian kernel.
*********************
*/
public void computeDensitiesGaussian() {
System.out.println("radius = " + radius);
densities = new double[dataset.numInstances()];
double tempDistance;
for (int i = 0; i < dataset.numInstances(); i++) {
for (int j = 0; j < dataset.numInstances(); j++) {
tempDistance = distance(i, j);
densities[i] += Math.exp(-tempDistance * tempDistance / radius / radius);
} // Of for j
} // Of for i
System.out.println("The densities are " + Arrays.toString(densities) + "\r\n");
}// Of computeDensitiesGaussian
/**
********************
* Compute the distance to its master.
*********************
*/
public void computeDistanceToMaster() {
distanceToMaster = new double[dataset.numInstances()];
masters = new int[dataset.numInstances()];
descendantDensities = new int[dataset.numInstances()];
instanceStatusArray = new int[dataset.numInstances()];
descendantDensities = mergeSortToIndices(densities);
distanceToMaster[descendantDensities[0]] = maximalDistance;
double tempDistance;
for (int i = 1; i < dataset.numInstances(); i++) {
// Initialize.
distanceToMaster[descendantDensities[i]] = maximalDistance;
for (int j = 0; j <= i - 1; j++) {
tempDistance = distance(descendantDensities[i], descendantDensities[j]);
if (distanceToMaster[descendantDensities[i]] > tempDistance) {
distanceToMaster[descendantDensities[i]] = tempDistance;
masters[descendantDensities[i]] = descendantDensities[j];
} // Of if
} // Of for j
} // Of for i
System.out.println("First compute, masters = " + Arrays.toString(masters));
System.out.println("descendantDensities = " + Arrays.toString(descendantDensities));
}// Of computeDistanceToMaster
/**
********************
* Compute priority. Element with higher priority is more likely to be selected
* as a cluster center. Now it is rho * distanceToMaster. It can also be
* rho^alpha * distanceToMaster.
*********************
*/
public void computePriority() {
priority = new double[dataset.numInstances()];
for (int i = 0; i < dataset.numInstances(); i++) {
priority[i] = densities[i] * distanceToMaster[i];
} // Of for i
}// Of computePriority
/**
********************
* Merge sort in descendant order to obtain an index array. The original array
* is unchanged. The method should be tested further. Examples: input [1.2, 2.3,
* 0.4, 0.5], output [1, 0, 3, 2]. input [3.1, 5.2, 6.3, 2.1, 4.4], output [2,
* 1, 4, 0, 3].
*
* @param paraArray the original array.
* @return The sorted indices.
*********************
*/
public static int[] mergeSortToIndices(double[] paraArray) {
int tempLength = paraArray.length;
int[][] resultMatrix = new int[2][tempLength];// For merge sort.
// Initialize
int tempIndex = 0;
for (int i = 0; i < tempLength; i++) {
resultMatrix[tempIndex][i] = i;
} // Of for i
// Merge
int tempCurrentLength = 1;
// The indices for current merged groups.
int tempFirstStart, tempSecondStart, tempSecondEnd;
while (tempCurrentLength < tempLength) {
// Divide into a number of groups.
// Here the boundary is adaptive to array length not equal to 2^k.
for (int i = 0; i < Math.ceil((tempLength + 0.0) / tempCurrentLength / 2); i++) {
// Boundaries of the group
tempFirstStart = i * tempCurrentLength * 2;
tempSecondStart = tempFirstStart + tempCurrentLength;
tempSecondEnd = tempSecondStart + tempCurrentLength - 1;
if (tempSecondEnd >= tempLength) {
tempSecondEnd = tempLength - 1;
} // Of if
// Merge this group
int tempFirstIndex = tempFirstStart;
int tempSecondIndex = tempSecondStart;
int tempCurrentIndex = tempFirstStart;
if (tempSecondStart >= tempLength) {
for (int j = tempFirstIndex; j < tempLength; j++) {
resultMatrix[(tempIndex + 1) % 2][tempCurrentIndex] = resultMatrix[tempIndex % 2][j];
tempFirstIndex++;
tempCurrentIndex++;
} // Of for j
break;
} // Of if
while ((tempFirstIndex <= tempSecondStart - 1) && (tempSecondIndex <= tempSecondEnd)) {
if (paraArray[resultMatrix[tempIndex % 2][tempFirstIndex]] >= paraArray[resultMatrix[tempIndex
% 2][tempSecondIndex]]) {
resultMatrix[(tempIndex + 1) % 2][tempCurrentIndex] = resultMatrix[tempIndex
% 2][tempFirstIndex];
tempFirstIndex++;
} else {
resultMatrix[(tempIndex + 1) % 2][tempCurrentIndex] = resultMatrix[tempIndex
% 2][tempSecondIndex];
tempSecondIndex++;
} // Of if
tempCurrentIndex++;
} // Of while
// Remaining part
for (int j = tempFirstIndex; j < tempSecondStart; j++) {
resultMatrix[(tempIndex + 1) % 2][tempCurrentIndex] = resultMatrix[tempIndex % 2][j];
tempCurrentIndex++;
} // Of for j
for (int j = tempSecondIndex; j <= tempSecondEnd; j++) {
resultMatrix[(tempIndex + 1) % 2][tempCurrentIndex] = resultMatrix[tempIndex % 2][j];
tempCurrentIndex++;
} // Of for j
} // Of for i
tempCurrentLength *= 2;
tempIndex++;
} // Of while
return resultMatrix[tempIndex % 2];
}// Of mergeSortToIndices
/**
********************
* Cluster based active learning.
*
* @param paraBlock The given block. This block must be sorted according to the
* priority in descendant order.
*********************
*/
public void clusterBasedActiveLearning(int[] paraBlock) {
System.out.println("clusterBasedAcrtiveLearning for block " + Arrays.toString(paraBlock));
// Step 1.How many labels are queried for this block.
int tempExpectedQueries = (int) Math.sqrt(paraBlock.length);
int tempNumQuery = 0;
for (int i = 0; i < paraBlock.length; i++) {
if (instanceStatusArray[paraBlock[i]] == 1) {
tempNumQuery++;
} // Of if
} // Of for i
// Step 2.Vote for small blocks.
if ((tempNumQuery >= tempExpectedQueries) && (paraBlock.length <= smallBlockThreshold)) {
System.out.println(
"" + tempNumQuery + " instances are queried, vote for block: \r\n" + Arrays.toString(paraBlock));
vote(paraBlock);
return;
} // Of if
// Step 3.Query enough labels.
for (int i = 0; i < tempExpectedQueries; i++) {
if (numQuery >= maxNumQuery) {
System.out.println("No more queries are provided, numQuery = " + numQuery + ".");
vote(paraBlock);
return;
} // Of if
if (instanceStatusArray[paraBlock[i]] == 0) {
instanceStatusArray[paraBlock[i]] = 1;
predictedLabels[paraBlock[i]] = (int) dataset.instance(paraBlock[i]).classValue();
numQuery++;
} // Of if
} // Of for i
// Step 4.Pure?
int tempFirstLabel = predictedLabels[paraBlock[0]];
boolean tempPure = true;
for (int i = 1; i < tempExpectedQueries; i++) {
if (predictedLabels[paraBlock[i]] != tempFirstLabel) {
tempPure = false;
break;
} // Of if
} // Of for i
if (tempPure) {
System.out.println("Classify for pure block: " + Arrays.toString(paraBlock));
for (int i = tempExpectedQueries; i < paraBlock.length; i++) {
if (instanceStatusArray[paraBlock[i]] == 0) {
predictedLabels[paraBlock[i]] = tempFirstLabel;
instanceStatusArray[paraBlock[i]] = 2;
} // Of if
} // Of for i
return;
} // Of if
// Step 5.Split in two and process them independently.
int[][] tempBlocks = clusterInTwo(paraBlock);
for (int i = 0; i < 2; i++) {
// Attention:recursive invoking here.
clusterBasedActiveLearning(tempBlocks[i]);
} // Of for i
}// Of clusterBasedActiveLearning
/**
********************
* Classify instances in the block by simple voting.
*
* @param paraBlock The given block.
*********************
*/
public void vote(int[] paraBlock) {
int[] tempClassCounts = new int[dataset.numClasses()];
for (int i = 0; i < paraBlock.length; i++) {
if (instanceStatusArray[paraBlock[i]] == 1) {
tempClassCounts[(int) dataset.instance(paraBlock[i]).classValue()]++;
} // Of if
} // Of for i
int tempMaxClass = -1;
int tempMaxCount = -1;
for (int i = 0; i < tempClassCounts.length; i++) {
if (tempMaxCount < tempClassCounts[i]) {
tempMaxClass = i;
tempMaxCount = tempClassCounts[i];
} // Of if
} // Of for i
// Classify unprocessed instances.
for (int i = 0; i < paraBlock.length; i++) {
if (instanceStatusArray[paraBlock[i]] == 0) {
predictedLabels[paraBlock[i]] = tempMaxClass;
instanceStatusArray[paraBlock[i]] = 2;
} // Of if
} // Of for i
}// Of vote
/**
********************
* Cluster a block in two. According to the master tree.
*
* @param paraBlock The given block.
* @return The new blocks where the two most represent instances serve as the
* root.
*********************
*/
public int[][] clusterInTwo(int[] paraBlock) {
// Reinitialize. In fact,only instances in the given block is considered.
Arrays.fill(clusterIndices, -1);
// Initialize the cluster number of the two roots.
for (int i = 0; i < 2; i++) {
clusterIndices[paraBlock[i]] = i;
} // Of for i
for (int i = 0; i < paraBlock.length; i++) {
if (clusterIndices[paraBlock[i]] != -1) {
// Already have a cluster number.
continue;
} // Of if
clusterIndices[paraBlock[i]] = coincideWithMaster(masters[paraBlock[i]]);
} // Of for i
// The sub blocks.
int[][] resultBlocks = new int[2][];
int tempFirstBlockCount = 0;
for (int i = 0; i < clusterIndices.length; i++) {
if (clusterIndices[i] == 0) {
tempFirstBlockCount++;
} // Of if
} // Of for i
resultBlocks[0] = new int[tempFirstBlockCount];
resultBlocks[1] = new int[paraBlock.length - tempFirstBlockCount];
// Copy. You can design shorter code when the number of cluster is greater than
// 2.
int tempFirstIndex = 0;
int tempSecondIndex = 0;
for (int i = 0; i < paraBlock.length; i++) {
if (clusterIndices[paraBlock[i]] == 0) {
resultBlocks[0][tempFirstIndex] = paraBlock[i];
tempFirstIndex++;
} else {
resultBlocks[1][tempSecondIndex] = paraBlock[i];
tempSecondIndex++;
} // Of if
} // Of for i
System.out.println("Split (" + paraBlock.length + ") instances " + Arrays.toString(paraBlock) + "\r\nto ("
+ resultBlocks[0].length + ") instances " + Arrays.toString(resultBlocks[0]) + "\r\nand ("
+ resultBlocks[1].length + ") instances " + Arrays.toString(resultBlocks[1]));
return resultBlocks;
}// Of clusterInTwo
/**
********************
* The block of a node should be same as its master. This recursive method is
* efficient.
*
* @param paraIndex The index of the given node.
* @return The cluster index of the current node.
*********************
*/
public int coincideWithMaster(int paraIndex) {
if (clusterIndices[paraIndex] == -1) {
int tempMaster = masters[paraIndex];
clusterIndices[paraIndex] = coincideWithMaster(tempMaster);
} // Of if
return clusterIndices[paraIndex];
}// Of coincideWithMaster
/**
*********************
* Show the statistics information.
*********************
*/
public String toString() {
int[] tempStatusCounts = new int[3];
double tempCorrect = 0;
for (int i = 0; i < dataset.numInstances(); i++) {
tempStatusCounts[instanceStatusArray[i]]++;
if (predictedLabels[i] == (int) dataset.instance(i).classValue()) {
tempCorrect++;
} // Of if
} // Of for i
String resultString = "(unhandled, queried, classified) = " + Arrays.toString(tempStatusCounts);
resultString += "\r\nCorrect = " + tempCorrect + ", accuracy = " + (tempCorrect / dataset.numInstances());
return resultString;
}// Of toString
/**
**********************************
* The entrance of the program.
*
* @param args: Not used now.
**********************************
*/
public static void main(String[] args) {
long tempStart = System.currentTimeMillis();
System.out.println("Starting ALEC.");
// String arffFilename = "F:/sampledataMain/iris.arff";
String arffFilename = "F:/sampledataMain/mushroom.arff";
Alec tempAlec = new Alec(arffFilename);
// The settings for iris
// tempAlec.clusterBasedActiveLearning(0.15, 30, 3);
// The settings for mushroom
tempAlec.clusterBasedActiveLearning(0.1, 800, 3);
System.out.println(tempAlec);
long tempEnd = System.currentTimeMillis();
System.out.println("Runtime: " + (tempEnd - tempStart) + "ms.");
}// Of main
}// Of class Alec
方法介绍:
public void computeMaximalDistance():用来求任意两个实例之间的最大距离;
public double distance(int paraI, int paraJ):求两个实例之间的欧式距离;
public void clusterBasedActiveLearning(double paraRatio, int paraMaxNumQuery, int paraSmallBlockThreshold):使数据主动学习的初始方法,在这个方法中主要设置了相关参数并获取重要性和距离等数据,数据的主动学习过程主要在它的重载方法public void clusterBasedActiveLearning(int[] paraBlock)中实现;
public void computeDensitiesGaussian():通过该方法可以计算出各个实例的重要性,当
d
e
n
s
i
t
i
e
s
[
i
]
densities[i]
densities[i]的值越大,说明实例
i
i
i附近的实例就越多,其重要性也越大。其中Math.exp(-tempDistance * tempDistance / radius / radius):(tempDistance * tempDistance / radius / radius)的结果一定
≥
0
\ge 0
≥0 ,可以很好避免整体计算结果越界,当
t
e
m
p
D
i
s
t
a
n
c
e
≤
r
a
d
i
u
s
tempDistance\le radius
tempDistance≤radius半径时,计算结果的区间为
[
1
e
,
1
]
[\frac{1}{e},1]
[e1,1],实例
j
j
j与实例
i
i
i的距离越远,
e
−
t
e
m
p
D
i
s
t
a
n
c
e
2
r
a
d
d
i
u
s
2
e^{-\frac{tempDistance^2}{raddius^2}}
e−raddius2tempDistance2值越小,也能说明实例
i
i
i代表实例
j
j
j的重要性越低。
public void computeDistanceToMaster():通过遍历各个实例之间的距离,获取实例到各自的monster的距离,构建monster树。由于实例
i
i
i到实例
j
j
j的距离与实例
j
j
j到实例
i
i
i的距离是相同的,所以只用计算一遍就可以了。每次会将新计算得到的距离与上次获得的最近距离进行比较(离当前实例越近,是当前实例的master的可能性就越大),当新的值更小时,就更新实例
i
i
i的master为当前的实例
j
j
j,并且更新其到master的距离。根据descendantDensities来构建,可以把密度最大的实例作为根结点实例;
public void computePriority():计算各个结点的代表性(重要性*距离);
public static int[] mergeSortToIndices(double[] paraArray):归并排序算法获取降序排序的序列索引;
public void vote(int[] paraBlock):用于选出某一个簇中个数最多标签,并将所有未查询的实例的标签设置为该标签。
public int[][] clusterInTwo(int[] paraBlock):将一个簇分成两个簇 ;
针对Iris数据的运行结果:

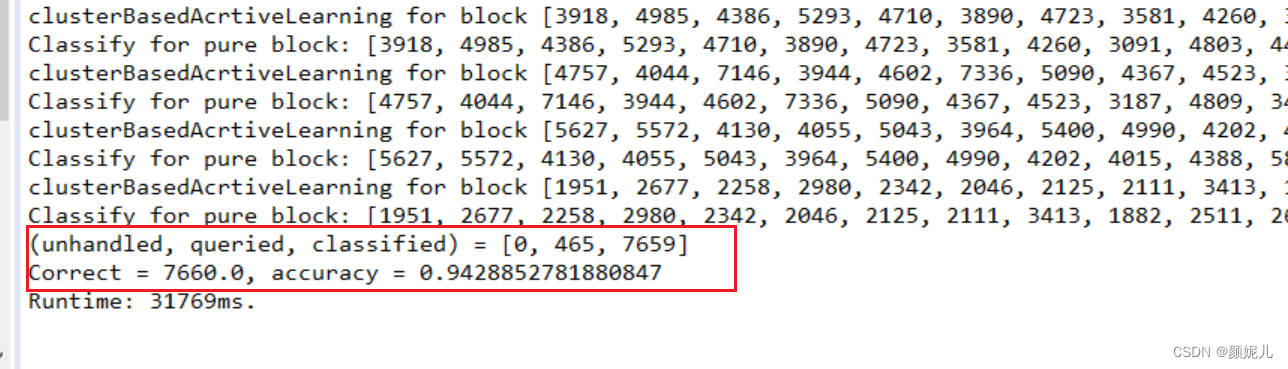
针对mushroom数据的运行结果:
这篇代码我看了两天,我真的是越看越不理解,先把问题记在这儿,等过几天再来看,思路可能会更清晰一点:
Q1:在clusterInTwo(int[] paraBlock)方法中初始化两个簇的根结点时,为什么直接默认初始前面两个实例?形参paraBlock对应的实参是descendantRepresentatives,该数组的前两个实例只能说明它们的代表性强但并没有表示它们的标签不同,我想的是在调用clusterInTwo前的判纯过程中获取到第一个与前面查询的标签不同的实例,将该这两个实例作为不同簇的根结点。我按照这个想法修改了代码,但效果没有之前的代码好,但我想了好久都没有想到原因🙃。
Q2:在clusterInTwo(int[] paraBlock)方法中对初始化根结点时,按理说是要保证必须把根据descendantDensities建立master树的根结点初始化掉的,才能保证后面的子结点通过其父结点找到它对应的簇,但是由于初始化时(按照老师的初始化代码)descendantRepresentatives[0]所对应的实例不一定与descendantDensitiesdescendantDensities[0](master树根结点的实例索引)指向同一个实例,所以按理说在coincideWithMaster过程中是有栈溢出的可能的吧?
我忽略了在computeDistanceToMaster方法中设置distanceToMaster[descendantDensities[0]] = maximalDistance,这样可以保证descendantDensities[0]与descendantRepresentatives[0]指向同一个实例对象,也就是一定能初始化到根元素。
Q3:在public void clusterBasedActiveLearning(int[] paraBlock) 方法中,
if ((tempNumQuery >= tempExpectedQueries) && (paraBlock.length <= smallBlockThreshold))
其中tempNumQuery 表示当前簇已经的查询个数,tempExpectedQueries表示期望当前簇能查询的个数,paraBlock.length表示当前簇的大小,smallBlockThreshold最小簇的阈值。只有当这两个条件都满足时会停止查询,但如果满足前面的条件,而不满足后面的条件,那么会继续查询而且是根据paraBlock从前往后依次查询的,但令我觉得矛盾的是判断当前簇是否为纯的时候,又只看tempExpectedQueries的数据,针对刚才提到的情况,那么查询是不是没有多大意义了。
就先这样吧,可能钻牛角尖了。
























 974
974











 被折叠的 条评论
为什么被折叠?
被折叠的 条评论
为什么被折叠?








