from ase import Atoms
from ase.optimize import BFGS
from ase.io.trajectory import Trajectory
from ase.constraints import FixInternals
from ase.constraints import FixAtoms
import math
from xtb.ase.calculator import XTB
import plotly.graph_objects as go
import pandas as pd
import numpy as np
sita = math.radians(107.23367921042804)
positions = [[3, 0, 0], [0, 0, 0], [3 * math.cos(sita), 3 * math.sin(sita), 0]]
water = Atoms('HOH', positions = positions)
angle_indices = [0, 1, 2]
angle = [water.get_angle(*angle_indices), angle_indices]
c1 = FixInternals(angles_deg=[angle])
water.set_constraint(c1)
c2 = FixAtoms(indices=[atom.index for atom in water if atom.symbol == 'O'])
water.set_constraint(c2)
water.calc = XTB(method = "GFN2-xTB")
opt = BFGS(water, trajectory = 'opt.traj')
opt.run(fmax = 0.05)
traj = Trajectory('opt.traj')
H1_O_d = []
H2_O_d = []
e_traj = []
for atoms in traj:
H1_O_d.append(atoms.get_distance(0, 1))
H2_O_d.append(atoms.get_distance(1, 2))
e_traj.append(atoms.get_potential_energy())
#print(atoms.positions, atoms.get_potential_energy(), atoms.get_angle(0, 1, 2))
H1 = np.linspace(0.5, 3, 100)
H2 = H1
e_surface = {}
for H1_position in H1:
e = []
for H2_position in H2:
water = Atoms('HOH', positions=[[H1_position, 0, 0], [0, 0, 0], [H2_position * np.cos(sita), H2_position * np.sin(sita), 0]])
water.calc = XTB(method = "GFN2-xTB")
e.append(water.get_potential_energy())
e_surface_temp = {H1_position: e}
e_surface.update(e_surface_temp)
e_surface_df = pd.DataFrame(e_surface)
fig = go.Figure(data=[go.Surface(z=e_surface_df, x=H1, y=H2)])
fig.update_traces(contours_z=dict(show=True, usecolormap=True, highlightcolor="limegreen", project_z=True, start=0, end=50, size=1))
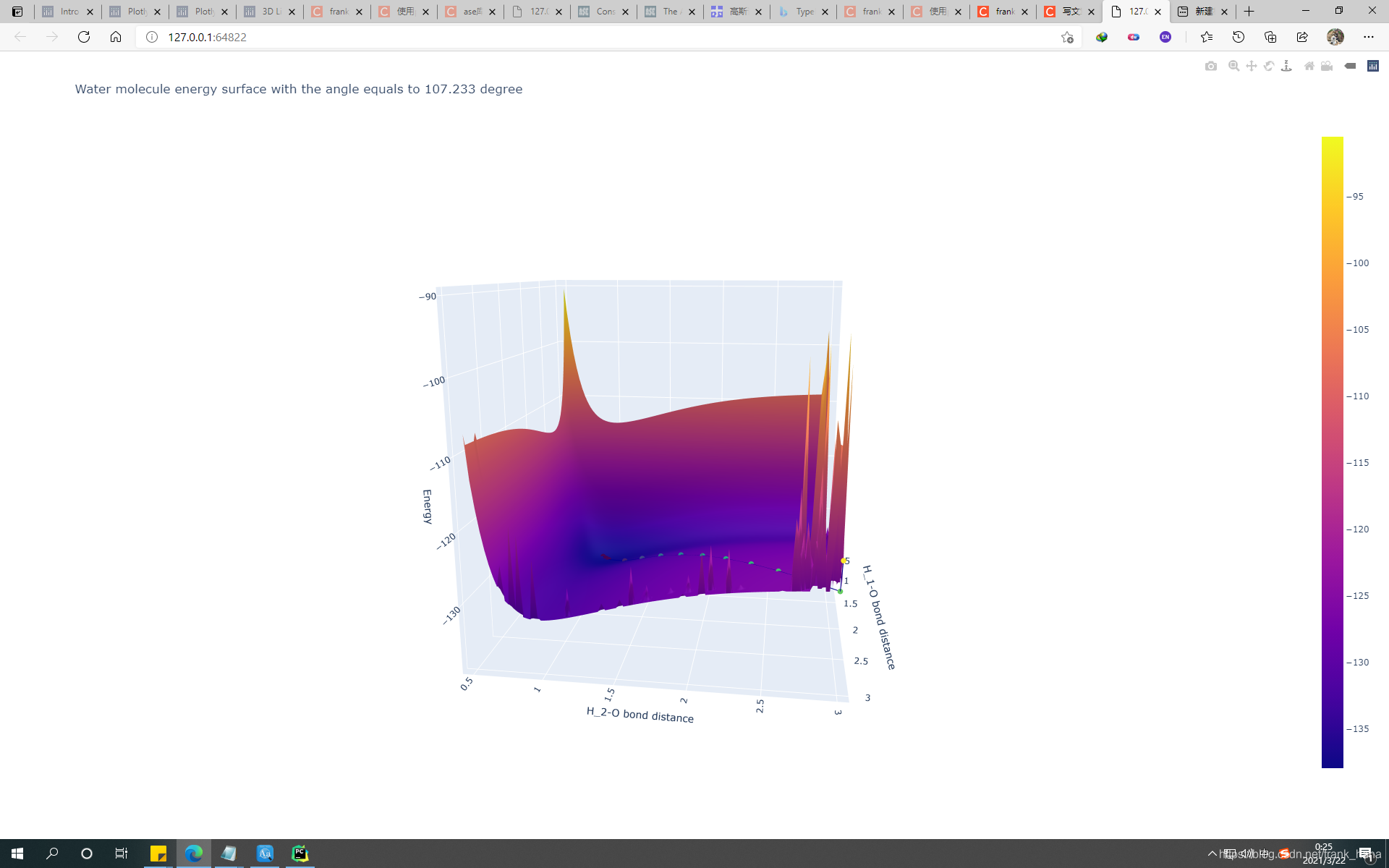
fig.update_layout(title="Water molecule energy surface with the angle equals to 107.233 degree", scene=dict(xaxis_title='H_1-O bond distance', yaxis_title='H_2-O bond distance', zaxis_title='Energy'))
fig.add_traces(data=go.Scatter3d(
x=H1_O_d, y=H2_O_d, z=e_traj,
marker=dict(
size=4, color=e_traj, colorscale='Viridis'
),
line=dict(
color='darkblue', width=2
)
))
fig.show()
结果如下:
放大:



H原子和O原子相距3埃时,出现尖峰,推测两个H-O键已经断开,半尖峰时只断一个键。
难点:
为了控制自由度,将O原子固定,H-O-H夹角固定。







 博客分析了H原子和O原子间距与H-O键断开的关系,当二者相距3埃时出现尖峰,此时两个H-O键已断开,半尖峰时只断一个键。还提到控制自由度的难点,需将O原子和H-O-H夹角固定。
博客分析了H原子和O原子间距与H-O键断开的关系,当二者相距3埃时出现尖峰,此时两个H-O键已断开,半尖峰时只断一个键。还提到控制自由度的难点,需将O原子和H-O-H夹角固定。

















 被折叠的 条评论
为什么被折叠?
被折叠的 条评论
为什么被折叠?








