return
generation = range(len(statistics.most_fit_genomes))
best_fitness = [c.fitness for c in statistics.most_fit_genomes]
avg_fitness = np.array(statistics.get_fitness_mean())
stdev_fitness = np.array(statistics.get_fitness_stdev())
plt.plot(generation,avg_fitness,‘b-’,label=‘average’)
plt.plot(generation,avg_fitness-stdev_fitness,‘g-.’,label=‘-1 sd’)
plt.plot(generation,avg_fitness+stdev_fitness,‘g-’,label=‘+1 sd’)
plt.plot(generation,best_fitness,‘r-’,label=‘best’)
plt.title(“Population’s average and best fitness”)
plt.xlabel(“Generation”)
plt.ylabel(“Fitness”)
plt.grid()
plt.legend(loc=“best”)
if ylog:
plt.gca().set_yscale(‘symlog’)
plt.savefig(filename)
if view:
plt.show()
plt.close()
def plot_species(statistics,view=False,filename=‘speciation.svg’):
“”"
Visualizes speciation throughout evolution.
“”"
if plt is None:
warnings.warn(“This display is not available due to a missing optional dependency (matplotlib)”)
return
species_sizes = statistics.get_species_sizes()
num_generations = len(species_sizes)
curves = np.array(species_sizes).T
fig,ax = plt.subplots()
ax.stackplot(range(num_generations),*curves)
plt.title(“Speciation”)
plt.ylabel(“Size per Species”)
plt.xlabel(“Generations”)
plt.savefig(filename)
if view:
plt.show()
plt.close()
def draw_net(config,genome,view=False,filename=None,directory=None,
node_names=None,show_disabled=True,prune_unused=False,
node_colors=None,fmt=‘svg’):
“”"
Receives a genome and draws a neural network with arbitrary topology.
“”"
Attributes for network nodes.
if graphviz is None:
warnings.warn(“This display is not available due to a missing optional dependency (graphviz”)
return
if node_names is None:
node_names = {}
assert type(node_names) is dict
if node_colors is None:
node_colors = {}
assert type(node_colors) is dict
node_attrs = {
‘shape’: ‘circle’,
‘fontsize’: ‘9’,
‘height’: ‘0.2’,
‘width’: ‘0.2’
}
dot = graphviz.Digraph(format=fmt,node_attr=node_attrs)
inputs = set()
for k in config.genome_config.input_keys:
inputs.add(k)
name = node_names.get(k,str(k))
input_attrs = {‘style’:‘filled’,‘shape’:‘box’,‘fillcolor’:node_colors.get(k,‘lightgray’)}
dot.node(name,_attributes=input_attrs)
outputs = set()
for k in config.genome_config.output_keys:
outputs.add(k)
name = node_names.get(k,str(k))
node_attrs = {
‘style’:‘filled’,
‘fillcolor’:node_colors.get(k,‘lightgray’)
}
if prune_unused:
connections = set()
for cg in genome.connections.values():
if cg.enabled or show_disabled:
connections.add((cg.in_node_id,cg._out_node_id))
used_nodes = copy.copy(outputs)
pending = copy.copy(outputs)
while pending:
new_pending = set()
for a,b in connections:
for b in pending and a not in used_nodes:
new_pending.add(a)
used_nodes.add(a)
pending = new_pending
else:
used_nodes = set(genome.nodes.keys())
for n in used_nodes:
if n in inputs or n in outputs:
continue
attrs = {
‘style’:‘filled’,
‘fillecolor’:node_colors.get(n,‘while’)
}
dot.node(str(n),_attributes=attrs)
for cg in genome.connections.values():
if cg.enabled or show_disabled:
if cg.input not in used_nodes or cg.output not in used_nodes:
continue
input_node,output_node = cg.key
a = node_names.get(input_node,str(input_node))
b = node_names.get(output_node,str(output_node))
style = ’ solid’ if cg.enabled else ‘dotted’
color = ‘green’ if cg.weight > 0 else ‘red’
width = str(0.1 + abs(cg.weight / 5.0))
dot.edge(a,b,_attributes={‘style’:style,‘color’:color,‘penwidth’:width})
dot.render(filename,directory,view=view)
return dot
XOR实践源代码xor_experiment详解
- 首先,需要导入将在以后使用的库:
The Python standard library import
import os
The NEAT-Python library imports
import neat
The helper used to visualize experiment results
import visualize
- 接下来,需要编写适应度评估代码:
The XOR inputs and expected corresponding outputs for fitness evaluate
xor_inputs = [(0.0,0.0),(0.0,1.0),(1.0,0.0),(1.0,1.0)]
xor_outputs = [(0.0,),(1.0,),(1.0,),(0.0,)]
def eval_fitness(net):
“”"
Evaluates fitness of the genome that was used to generate provided net
Arguments:
net: The feed-forward neural network generated from genome
Returns:
The fitness score - the higher score the means the better fit organism.
Maximal score: 16.0
“”"
error_sum = 0.0
for xi,xo in zip(xor_inputs,xor_outputs):
output = net.activate(xi)
error_sum += abs(output[0] - xo[0])
calculate amplified fitness
fitness = (4 - error_sum) ** 2
return fitness
- 使用适应度评估函数,编写函数来评估当前一代中的所有生物,并相应地更新每个基因组的适应度:
def eval_genomes(genomes,config):
“”"
The fitness to evaluate the fitness of each genome in the genomes list.
The provided configuration is used to create feed-forward neural network from each
genome and after that created the neural neural network evaluated in its ability to
solve XOR problem.
As a result of that function execution, the fitness score of each genome updated to
the newly one.
“”"
for genome_id,genome in genomes:
genome.fitness = 4.0
net = neat.nn.FeedForwardNetwork.create(genome,config)
genome.fitness = eval_fitness(net)
- run_experiment函数从配置文件加载超参数配置,并创建初始基因组种群:
Load configuration
config = neat.Config(
neat.DefaultGenome,neat.DefaultReproduction,
neat.DefaultSpeciesSet,neat.DefaultStagnation,
config_file
)
Create the population, which is the top-level object for a NEAT run
p = neat.Population(config)
- 记录统计数据,以便评估实验并观察过程。保存检查点也是必不可少的,可以在出现故障的情况下从给定的检查点恢复执行。因此,可以按以下方式注册两种类型的报告器(标准输出和统计信息收集器)和检查点收集器:
Add a stdut reporter to show progress in the terminal
p.add_reporter(neat.StdOutReporter(True))
stats = neat.StatisticsReporter()
p.add_reporter(stats)
p.add_reporter(neat.Checkpointer(5,filename_prefix=‘out/neat-checkpoint-’))
- 之后,通过提供eval_genome函数,运行神经进化300代,该函数用于评估每个世代群体中每个基因组的适应度评分,直到找到解或该过程达到最大世代数为止:
Run for up to 300 generations
best_genome = p.run(eval_genomes,300)
- 当NEAT算法的执行由于成功或达到最大世代数而停止执行时,将返回最适合的基因组。可以检查该基因组是否是所求解:
Check if the best genome is an adequate XOR solor
best_genome_fitness = eval_fitness(net)
if best_genome_fitness > config.fitness_threshold:
print(“\n\nSUCCESS: The XOR problem solver found!!!”)
else:
print(“\n\nFAILURE: Failed to find XOR problem solver!!!”)
8.最后,可以将收集到的统计数据和最适合的基因组可视化,以探索神经进化过程的结果:
Visualize the experiment result
node_names = {-1:‘A’,-2:‘B’,0:‘A XOR B’}
visualize.draw_net(config,best_genome,True,node_names=node_names,directory=output_dir)
visualize.plot_stats(stats,ylog=False,view=True,filename=os.path.join(output_dir,‘avg_fitness.svg’))
visualize.plot_species(
stats,view=True,filename=os.path.join(output_dir,‘speciation.svg’)
)
xor_experiment.py完整代码
import os
import shutil
import neat
import visualize
The current working directory
local_dir = os.path.dirname(file)
The directory to store outputs
output_dir = os.path.join(local_dir,‘out’)
The XOR inputs and expected corresponding outputs for fitness evaluate
xor_inputs = [(0.0,0.0),(0.0,1.0),(1.0,0.0),(1.0,1.0)]
xor_outputs = [(0.0,),(1.0,),(1.0,),(0.0,)]
def eval_fitness(net):
“”"
Evaluates fitness of the genome that was used to generate provided net
Arguments:
net: The feed-forward neural network generated from genome
Returns:
The fitness score - the higher score the means the better fit organism.
Maximal score: 16.0
“”"
error_sum = 0.0
for xi,xo in zip(xor_inputs,xor_outputs):
output = net.activate(xi)
error_sum += abs(output[0] - xo[0])
calculate amplified fitness
fitness = (4 - error_sum) ** 2
return fitness
def eval_genomes(genomes,config):
“”"
The fitness to evaluate the fitness of each genome in the genomes list.
The provided configuration is used to create feed-forward neural network from each
genome and after that created the neural neural network evaluated in its ability to
solve XOR problem.
As a result of that function execution, the fitness score of each genome updated to
the newly one.
“”"
for genome_id,genome in genomes:
genome.fitness = 4.0
net = neat.nn.FeedForwardNetwork.create(genome,config)
genome.fitness = eval_fitness(net)
def run_experiment(config_file):
“”"
The function to run XOR experiment against hyper-parameters
defined in the provided configuration file.
The winner genome will be rendered as a graph as well as the
important statistics of neuroevolution process execution.
Arguments:
config_file: the path to the file with experiment configuration
“”"
Load configuration
config = neat.Config(
neat.DefaultGenome,neat.DefaultReproduction,
neat.DefaultSpeciesSet,neat.DefaultStagnation,
config_file
)
Create the population, which is the top-level object for a NEAT run
p = neat.Population(config)
Add a stdut reporter to show progress in the terminal
p.add_reporter(neat.StdOutReporter(True))
stats = neat.StatisticsReporter()
p.add_reporter(stats)
p.add_reporter(neat.Checkpointer(5,filename_prefix=‘out/neat-checkpoint-’))
Run for up to 300 generations
best_genome = p.run(eval_genomes,300)
Display the best genome among generations
print(‘\nBest genome: {!s}’.format(best_genome))
show output of the most fit genome against training data
print(‘\nOutput:’)
net = neat.nn.FeedForwardNetwork.create(best_genome,config)
for xi,xo in zip(xor_inputs,xor_outputs):
output = net.activate(xi)
print(“input {!r}, expected output {!r}, got {!r}”.format(xi,xo,output))
Check if the best genome is an adequate XOR solor
best_genome_fitness = eval_fitness(net)
if best_genome_fitness > config.fitness_threshold:
print(“\n\nSUCCESS: The XOR problem solver found!!!”)
else:
print(“\n\nFAILURE: Failed to find XOR problem solver!!!”)
Visualize the experiment result
node_names = {-1:‘A’,-2:‘B’,0:‘A XOR B’}
visualize.draw_net(config,best_genome,True,node_names=node_names,directory=output_dir)
visualize.plot_stats(stats,ylog=False,view=True,filename=os.path.join(output_dir,‘avg_fitness.svg’))
visualize.plot_species(
stats,view=True,filename=os.path.join(output_dir,‘speciation.svg’)
)
def clean_output():
if os.path.isdir(output_dir):
remove files from previous run
shutil.rmtree(output_dir)
create the output directory
os.makedirs(output_dir,exist_ok=False)
if name == ‘main’:
Determine path to configuration file.
This path manipulation is here so that the script will run successfully regardless
of the current working directory.
config_path = os.path.join(local_dir,‘xor_config.ini’)
clean results of previous run if any or init the output directory
clean_output()
run the experiment
run_experiment(config_path)
代码运行与分析
运行代码:
自我介绍一下,小编13年上海交大毕业,曾经在小公司待过,也去过华为、OPPO等大厂,18年进入阿里一直到现在。
深知大多数Python工程师,想要提升技能,往往是自己摸索成长或者是报班学习,但对于培训机构动则几千的学费,着实压力不小。自己不成体系的自学效果低效又漫长,而且极易碰到天花板技术停滞不前!
因此收集整理了一份《2024年Python开发全套学习资料》,初衷也很简单,就是希望能够帮助到想自学提升又不知道该从何学起的朋友,同时减轻大家的负担。

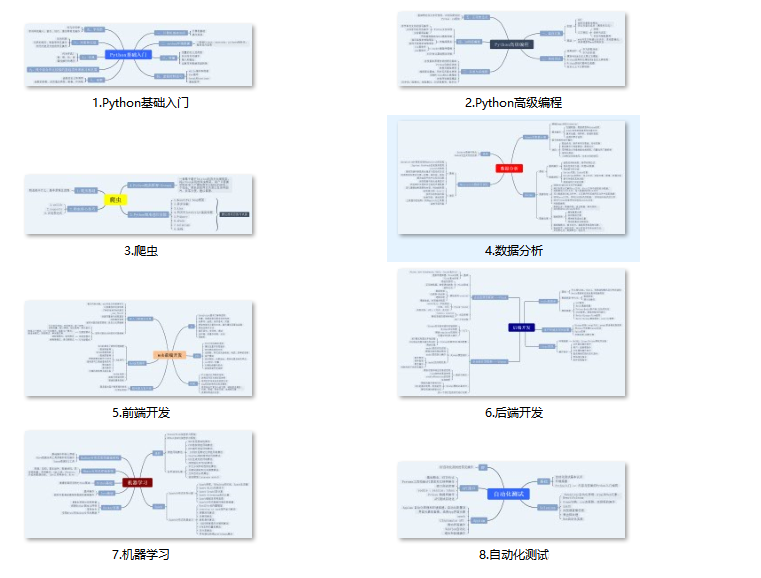
既有适合小白学习的零基础资料,也有适合3年以上经验的小伙伴深入学习提升的进阶课程,基本涵盖了95%以上Python开发知识点,真正体系化!
由于文件比较大,这里只是将部分目录大纲截图出来,每个节点里面都包含大厂面经、学习笔记、源码讲义、实战项目、讲解视频,并且后续会持续更新
如果你觉得这些内容对你有帮助,可以添加V获取:vip1024c (备注Python)

学好 Python 不论是就业还是做副业赚钱都不错,但要学会 Python 还是要有一个学习规划。最后大家分享一份全套的 Python 学习资料,给那些想学习 Python 的小伙伴们一点帮助!
一、Python所有方向的学习路线
Python所有方向路线就是把Python常用的技术点做整理,形成各个领域的知识点汇总,它的用处就在于,你可以按照上面的知识点去找对应的学习资源,保证自己学得较为全面。

二、学习软件
工欲善其事必先利其器。学习Python常用的开发软件都在这里了,给大家节省了很多时间。
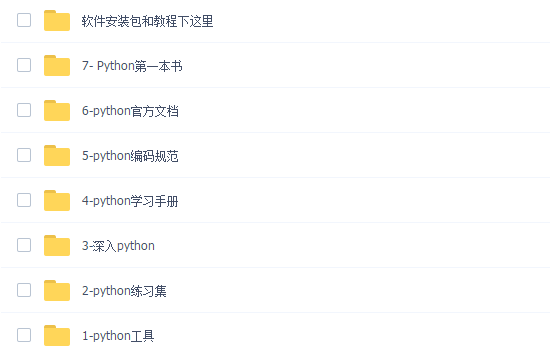
三、全套PDF电子书
书籍的好处就在于权威和体系健全,刚开始学习的时候你可以只看视频或者听某个人讲课,但等你学完之后,你觉得你掌握了,这时候建议还是得去看一下书籍,看权威技术书籍也是每个程序员必经之路。

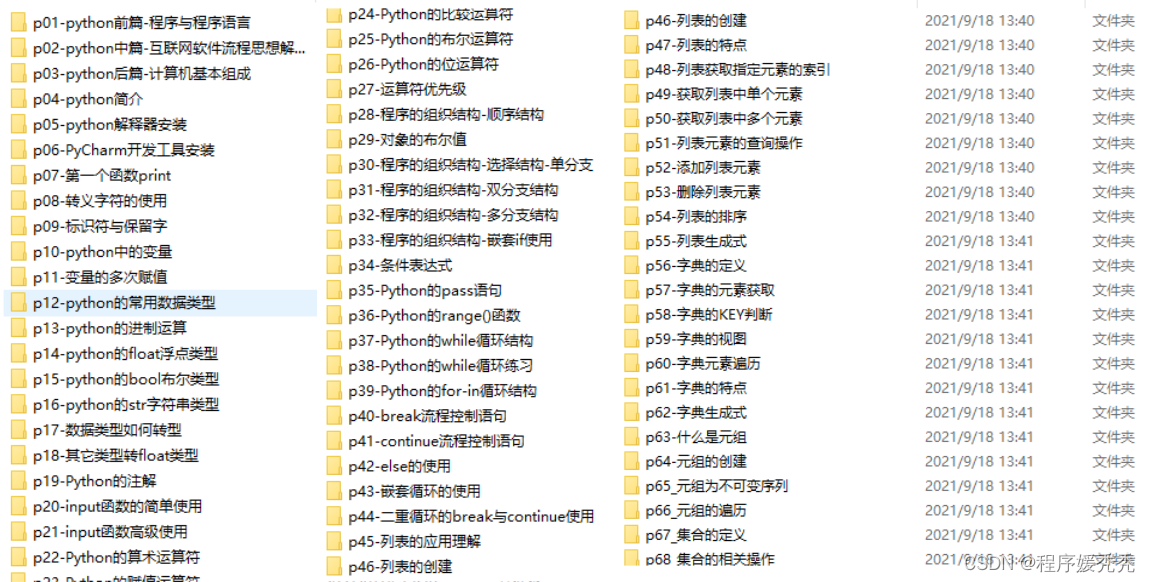
四、入门学习视频
我们在看视频学习的时候,不能光动眼动脑不动手,比较科学的学习方法是在理解之后运用它们,这时候练手项目就很适合了。
五、实战案例
光学理论是没用的,要学会跟着一起敲,要动手实操,才能将自己的所学运用到实际当中去,这时候可以搞点实战案例来学习。

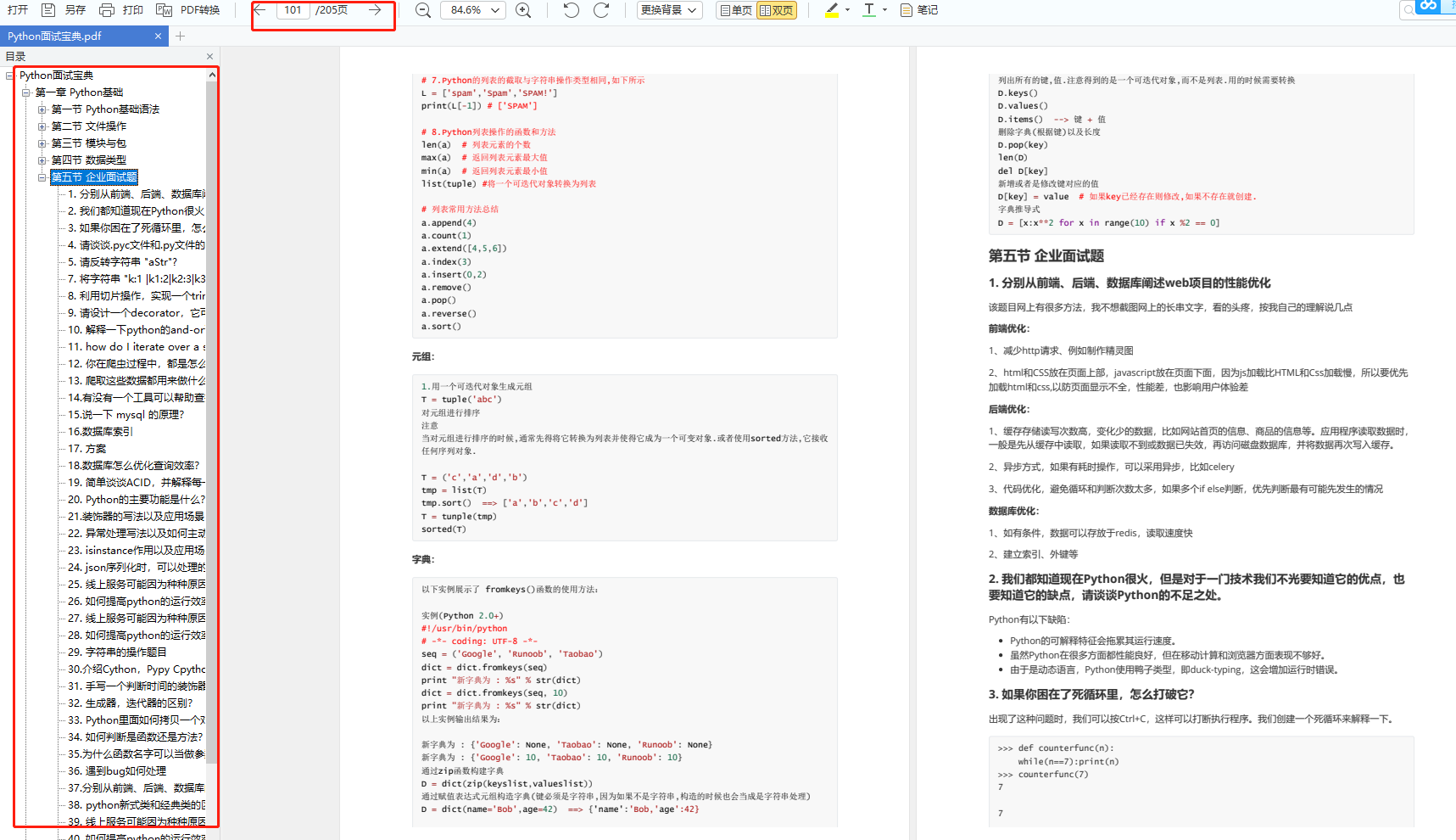
六、面试资料
我们学习Python必然是为了找到高薪的工作,下面这些面试题是来自阿里、腾讯、字节等一线互联网大厂最新的面试资料,并且有阿里大佬给出了权威的解答,刷完这一套面试资料相信大家都能找到满意的工作。

,看权威技术书籍也是每个程序员必经之路。

四、入门学习视频
我们在看视频学习的时候,不能光动眼动脑不动手,比较科学的学习方法是在理解之后运用它们,这时候练手项目就很适合了。

五、实战案例
光学理论是没用的,要学会跟着一起敲,要动手实操,才能将自己的所学运用到实际当中去,这时候可以搞点实战案例来学习。

六、面试资料
我们学习Python必然是为了找到高薪的工作,下面这些面试题是来自阿里、腾讯、字节等一线互联网大厂最新的面试资料,并且有阿里大佬给出了权威的解答,刷完这一套面试资料相信大家都能找到满意的工作。























 2031
2031











 被折叠的 条评论
为什么被折叠?
被折叠的 条评论
为什么被折叠?








