vec <- 1
names(vec) <- c("hsa:51343")
new_g <- g |> mutate(num=node_numeric(vec))
new_g
#> # A tbl_graph: 134 nodes and 157 edges
#> #
#> # A directed acyclic multigraph with 40 components
#> #
#> # A tibble: 134 × 23
#> name type reaction graphics_name x y width
#> <chr> <chr> <chr> <chr> <dbl> <dbl> <dbl>
#> 1 hsa:1029 gene <NA> CDKN2A, ARF,… 532 -218 46
#> 2 hsa:51343 gene <NA> FZR1, CDC20C… 981 -630 46
#> 3 hsa:4171 h… gene <NA> MCM2, BM28, … 553 -681 46
#> 4 hsa:23594 … gene <NA> ORC6, ORC6L.… 494 -681 46
#> 5 hsa:10393 … gene <NA> ANAPC10, APC… 981 -392 46
#> 6 hsa:10393 … gene <NA> ANAPC10, APC… 981 -613 46
#> # ℹ 128 more rows
#> # ℹ 16 more variables: height <dbl>, fgcolor <chr>,
#> # bgcolor <chr>, graphics_type <chr>, coords <chr>,
#> # xmin <dbl>, xmax <dbl>, ymin <dbl>, ymax <dbl>,
#> # orig.id <chr>, pathway_id <chr>, deseq2 <dbl>,
#> # padj <dbl>, converted_name <chr>, lfc <dbl>, num <dbl>
#> #
#> # A tibble: 157 × 6
#> from to type subtype_name subtype_value pathway_id
#> <int> <int> <chr> <chr> <chr> <chr>
#> 1 118 39 GErel expression --> hsa04110
#> 2 50 61 PPrel inhibition --| hsa04110
#> 3 50 61 PPrel phosphorylation +p hsa04110
#> # ℹ 154 more rows
将矩阵积分到tbl_graph
如果要在图形中反映表达式矩阵,则 和 函数可能很有用。通过指定基质和基因 ID,您可以将每个样品的数值分配给 . 分配由边连接的两个节点的总和,忽略组节点(Adnan 等人,2020 年)。edge_matrix``node_matrix``tbl_graph``edge_matrix
mat <- assay(vst(res))
new_g <- g |> edge_matrix(mat) |> node_matrix(mat)
new_g
#> # A tbl_graph: 134 nodes and 157 edges
#> #
#> # A directed acyclic multigraph with 40 components
#> #
#> # A tibble: 134 × 48
#> name type reaction graphics_name x y width
#> <chr> <chr> <chr> <chr> <dbl> <dbl> <dbl>
#> 1 hsa:1029 gene <NA> CDKN2A, ARF,… 532 -218 46
#> 2 hsa:51343 gene <NA> FZR1, CDC20C… 981 -630 46
#> 3 hsa:4171 h… gene <NA> MCM2, BM28, … 553 -681 46
#> 4 hsa:23594 … gene <NA> ORC6, ORC6L.… 494 -681 46
#> 5 hsa:10393 … gene <NA> ANAPC10, APC… 981 -392 46
#> 6 hsa:10393 … gene <NA> ANAPC10, APC… 981 -613 46
#> # ℹ 128 more rows
#> # ℹ 41 more variables: height <dbl>, fgcolor <chr>,
#> # bgcolor <chr>, graphics_type <chr>, coords <chr>,
#> # xmin <dbl>, xmax <dbl>, ymin <dbl>, ymax <dbl>,
#> # orig.id <chr>, pathway_id <chr>, deseq2 <dbl>,
#> # padj <dbl>, converted_name <chr>, lfc <dbl>,
#> # SRR14509882 <dbl>, SRR14509883 <dbl>, …
#> #
#> # A tibble: 157 × 34
#> from to type subtype_name subtype_value pathway_id
#> <int> <int> <chr> <chr> <chr> <chr>
#> 1 118 39 GErel expression --> hsa04110
#> 2 50 61 PPrel inhibition --| hsa04110
#> 3 50 61 PPrel phosphorylation +p hsa04110
#> # ℹ 154 more rows
#> # ℹ 28 more variables: from_nd <chr>, to_nd <chr>,
#> # SRR14509882 <dbl>, SRR14509883 <dbl>,
#> # SRR14509884 <dbl>, SRR14509885 <dbl>,
#> # SRR14509886 <dbl>, SRR14509887 <dbl>,
#> # SRR14509888 <dbl>, SRR14509889 <dbl>,
#> # SRR14509890 <dbl>, SRR14509891 <dbl>, …
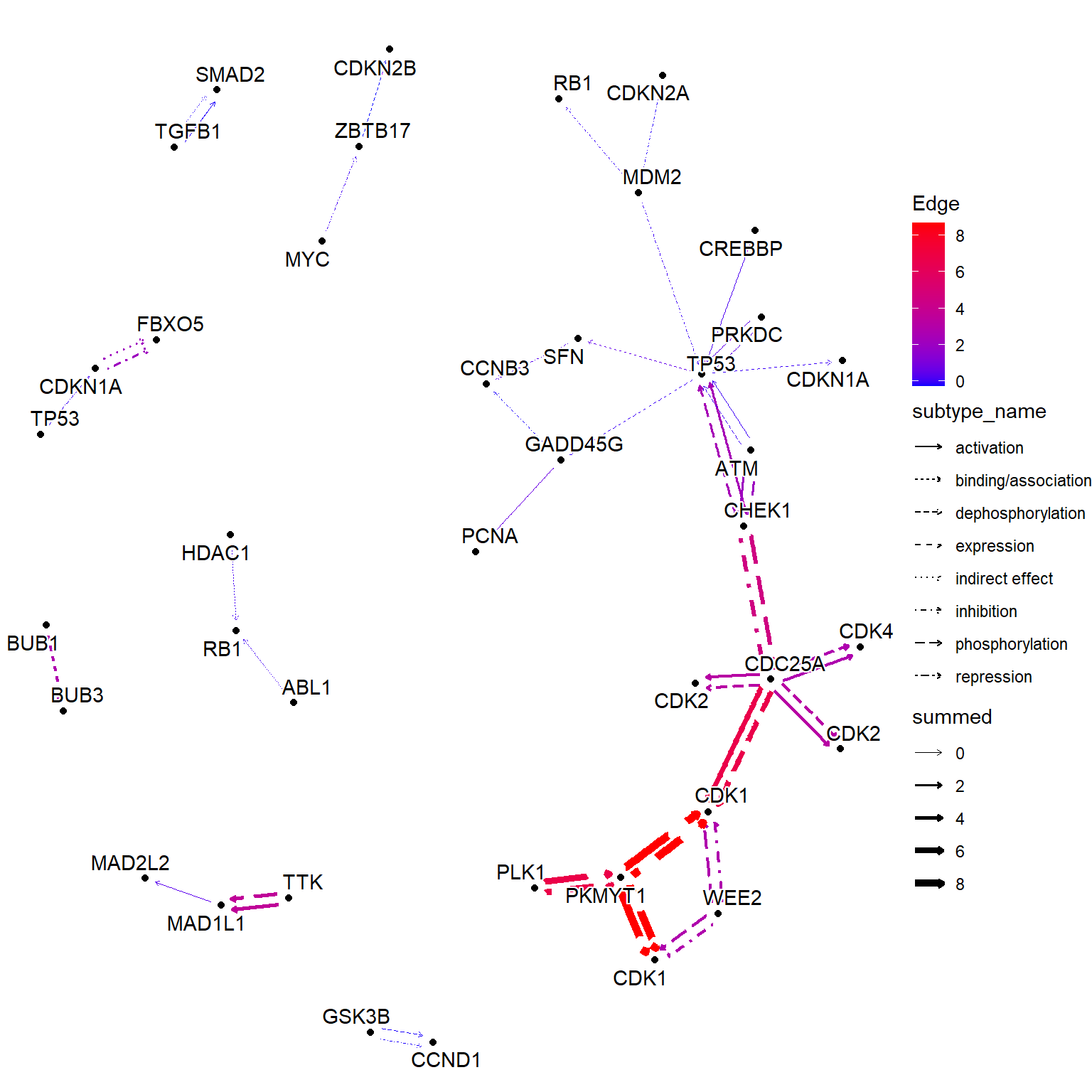
边值
相同的效果可以通过 获得,使用命名数值向量作为输入。此函数根据节点值添加边值。以下示例显示了将 LFC 组合到边缘。这与 的行为不同。edge_matrix``edge_numeric_sum``edge_numeric
## Numeric vector (name is SYMBOL)
vinflfc <- vinf$log2FoldChange |> setNames(row.names(vinf))
g |>
## Use graphics_name to merge
mutate(grname=strsplit(graphics_name, ",") |> vapply("[", 1, FUN.VALUE="a")) |>
activate(edges) |>
mutate(summed = edge_numeric_sum(vinflfc, name="grname")) |>
filter(!is.na(summed)) |>
activate(nodes) |>
mutate(x=NULL, y=NULL, deg=centrality_degree(mode="all")) |>
filter(deg>0) |>
ggraph(layout="nicely")+
geom_edge_parallel(aes(color=summed, width=summed,
linetype=subtype_name),
arrow=arrow(length=unit(1,"mm")),
start_cap=circle(2,"mm"),
end_cap=circle(2,"mm"))+
geom_node_point(aes(fill=I(bgcolor)))+
geom_node_text(aes(label=grname,
filter=type=="gene"),
repel=TRUE, bg.colour="white")+
scale_edge_width(range=c(0.1,2))+
scale_edge_color_gradient(low="blue", high="red", name="Edge")+
theme_void()
可视化多重富集结果
您可以可视化多个富集分析的结果。与将函数与类一起使用类似,可以在函数中使用一个函数。通过向此功能提供对象,如果结果中存在可视化的通路,则通路内的基因信息可以反映在图中。在这个例子中,除了上面提到的尿路上皮细胞的变化外,还比较了肾近端肾小管上皮细胞的变化(Assetta等人,2016)。ggkegg``enrichResult``append_cp``mutate``enrichResult
## These are RDAs storing DEGs
load("degListRPTEC.rda")
load("degURO.rda")
library(org.Hs.eg.db);
library(clusterProfiler);
input_uro <- bitr(uroUp, ## DEGs in urothelial cells
fromType = "SYMBOL",
toType = "ENTREZID",
OrgDb = org.Hs.eg.db)$ENTREZID
input_rptec <- bitr(gls$day3_up_rptec, ## DEGs at 3 days post infection in RPTECs
fromType = "SYMBOL",
toType = "ENTREZID",
OrgDb = org.Hs.eg.db)$ENTREZID
ekuro <- enrichKEGG(gene = input_uro)
ekrptec <- enrichKEGG(gene = input_rptec)
g1 <- pathway("hsa04110") |> mutate(uro=append_cp(ekuro, how="all"),
rptec=append_cp(ekrptec, how="all"),
converted_name=convert_id("hsa"))
ggraph(g1, layout="manual", x=x, y=y) +
geom_edge_parallel(width=0.5, arrow = arrow(length = unit(1, 'mm')),
start_cap = square(1, 'cm&







 最低0.47元/天 解锁文章
最低0.47元/天 解锁文章
















 被折叠的 条评论
为什么被折叠?
被折叠的 条评论
为什么被折叠?








