官方网址
https://omictools.com/get-homologues-tool
帮助文档
https://github.com/eead-csic-compbio/get_homologues/blob/master/manual_get_homologues.pdf
软件安装步骤
1 软件获取
wget https://github-cloud.s3.amazonaws.com/releases/46931871/af7a24f0-240a-11e6-8c41-e87b0ee2ed8d.tgz?X-Amz-Algorithm=AWS4-HMAC-SHA256&X-Amz-Credential=AKIAISTNZFOVBIJMK3TQ%2F20160622%2Fus-east-1%2Fs3%2Faws4_request&X-Amz-Date=20160622T142013Z&X-Amz-Expires=300&X-Amz-Signature=672fd831dd3147c8bbb843b5d1dbd2de9b7144cd5756069f48342bf69a4d5170&X-Amz-SignedHeaders=host&actor_id=0&response-content-disposition=attachment%3B%20filename%3Dget_homologues-x86_64-20160527.tgz&response-content-type=application%2Foctet-stream
2 解压
tar zxvf get_homologues-2.0.16.tar.gz
3 阅读README文件
注意:所有依赖软件均需安装至’get_homologues-2.0.16/bin’目录下。
依赖软件
1 COGtriangles(COGsoft)
2 hmmer(hmmer-3.1b2)
3 MCL(mcl-14-137)
4 BLAST(ncbi-blast-2.2.27+)
4 修改依赖软件文件夹名称
mv COGsoft.04.19.2012/ COGsoft
mv hmmer-3.1b2-linux-intel-x86_64/ hmmer-3.1b2
mv mcl-14-137* mcl-14-137
mv ncbi-blast-2.2.27+*/ ncbi-blast-2.2.27+
5 安装get_homologues
cd get_homologues-2.0.16
perl install.pl
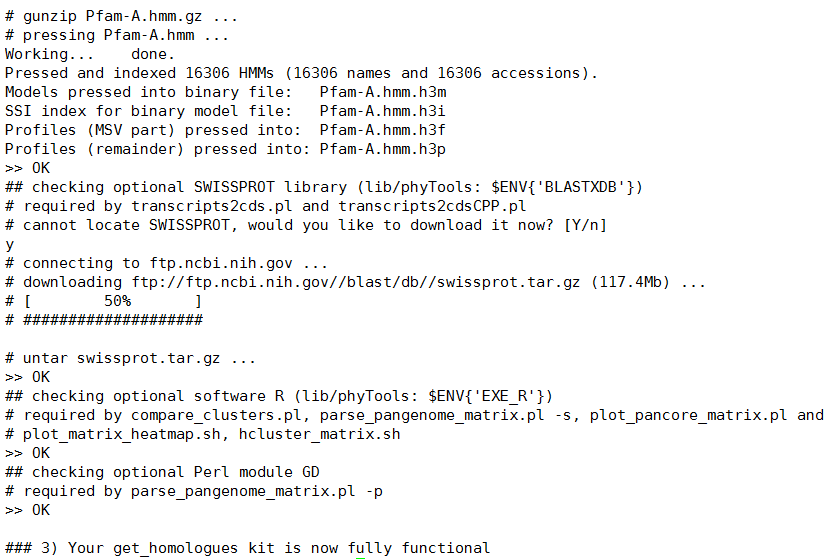
安装截图:

安装过程中会提示下载两个依赖软件(Pfam-A,SWISSPROT),选择‘y’即可。
























 5万+
5万+

 被折叠的 条评论
为什么被折叠?
被折叠的 条评论
为什么被折叠?








