import parallelproj
from copy import copy
import SimpleITK as sitk
import matplotlib.pyplot as plt
import array_api_compat.numpy as np
import array_api_compat.torch as xp
from array_api_strict._array_object import Array
def em_update(
x_cur: Array,
data: Array,
op: parallelproj.LinearOperator,
adjoint_ones: Array,
) -> Array:
"""EM update
Parameters
----------
x_cur : Array
current solution
data : Array
data
op : parallelproj.LinearOperator
linear forward operator
s : Array
contamination
adjoint_ones : Array
adjoint of ones
Returns
-------
Array
"""
epsilon = 1e-10
ybar = op(x_cur) + epsilon
return x_cur * op.adjoint(data / ybar) / adjoint_ones
dev = "cuda"
num_rings = 36
scanner = parallelproj.RegularPolygonPETScannerGeometry(
xp,
dev,
radius=120.0,
num_sides=16, # rsector
num_lor_endpoints_per_side=12, # trans crystal per sector
lor_spacing=3.25, # crystal spacing
ring_positions=xp.linspace(-50, 50, num_rings),
symmetry_axis=1,
)
lor_desc = parallelproj.RegularPolygonPETLORDescriptor(
scanner,
radial_trim=10,
max_ring_difference=None,
sinogram_order=parallelproj.SinogramSpatialAxisOrder.RVP,
)
vox = (1, 1, 1)
shape = (149, 95, 149)
proj = parallelproj.RegularPolygonPETProjector(
lor_desc, img_shape=shape, voxel_size=vox
)
hoffman = np.array(sitk.GetArrayFromImage(
sitk.ReadImage('/share/home/hannah/workStation/pytomography/parallelproj/hoffman.nii'))).astype(np.float32)
x = hoffman.transpose([1, 0, 2])
x = xp.asarray(x, device=dev) # 转到cuda中
x_fwd = proj(x) # 得到前向投影
# setup an attenuation image
x_att = 0.01 * xp.astype(x > 0, xp.float32)
# calculate the attenuation sinogram
att_sino = xp.exp(-proj(x_att))
att_op = parallelproj.ElementwiseMultiplicationOperator(att_sino)
res_model = parallelproj.GaussianFilterOperator(
proj.in_shape, sigma=4.5 / (2.35 * proj.voxel_size)
)
pet_lin_op = parallelproj.CompositeLinearOperator((att_op, proj, res_model))
num_subsets = 10
subset_views, subset_slices = proj.lor_descriptor.get_distributed_views_and_slices(
num_subsets, len(proj.out_shape)
)
_, subset_slices_non_tof = proj.lor_descriptor.get_distributed_views_and_slices(
num_subsets, 3
)
proj.clear_cached_lor_endpoints()
pet_subset_linop_seq = []
for i in range(num_subsets):
print(f"subset {i:02} containing views {subset_views[i]}")
# make a copy of the full projector and reset the views to project
subset_proj = copy(proj)
subset_proj.views = subset_views[i]
subset_att_op = parallelproj.ElementwiseMultiplicationOperator(
att_sino[subset_slices_non_tof[i]]
)
# add the resolution model and multiplication with a subset of the attenuation sinogram
pet_subset_linop_seq.append(
parallelproj.CompositeLinearOperator(
[
subset_att_op,
subset_proj,
res_model,
]
)
)
pet_subset_linop_seq = parallelproj.LinearOperatorSequence(pet_subset_linop_seq)
print("# ============================= OSEM =====================================")
# number of OSEM iterations
num_iter = 20 // len(pet_subset_linop_seq)
x = xp.ones(pet_lin_op.in_shape, dtype=xp.float64, device=dev)
y = x_fwd
# calculate A_k^H 1 for all subsets k
subset_adjoint_ones = [
x.adjoint(xp.ones(x.out_shape, dtype=xp.float64, device=dev))
for x in pet_subset_linop_seq
]
# OSEM iterations
for i in range(num_iter):
for k, sl in enumerate(subset_slices):
print(f"OSEM iteration {(k + 1):03} / {(i + 1):03} / {num_iter:03}", end="\r")
x = em_update(
x, y[sl], pet_subset_linop_seq[k], subset_adjoint_ones[k]
)
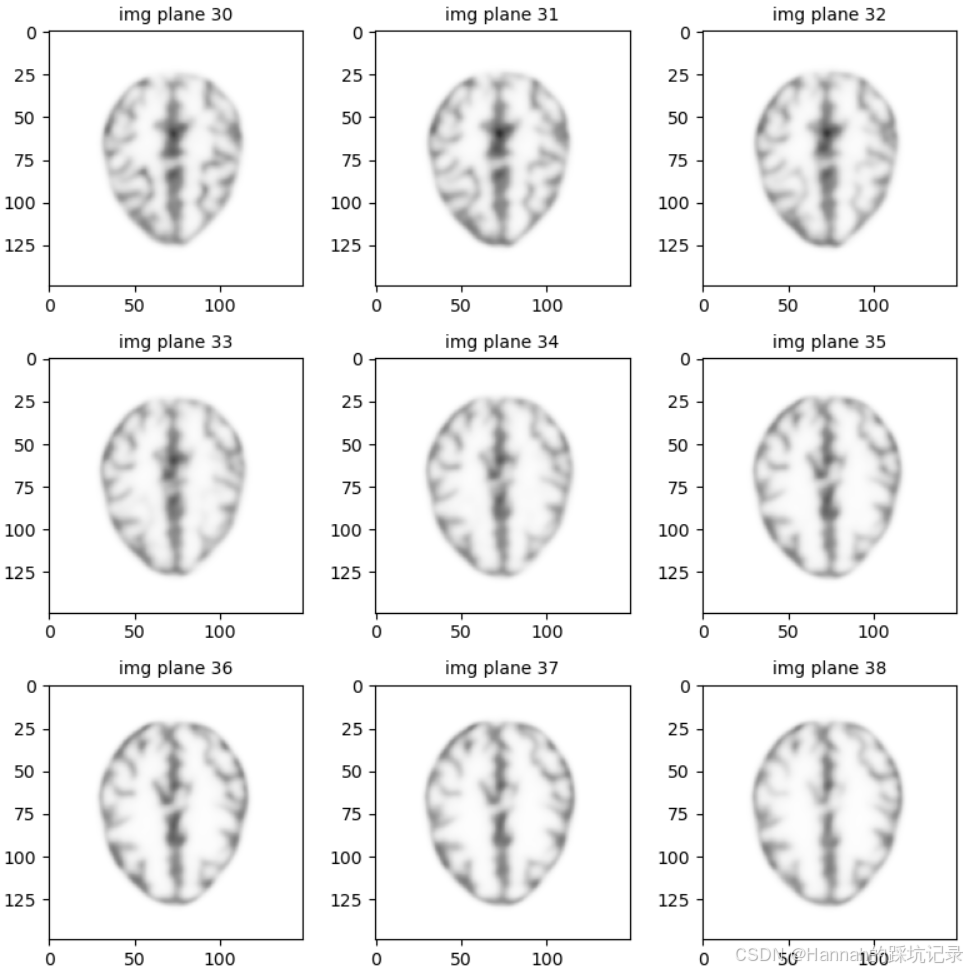
slice = 30
fig5, ax5 = plt.subplots(3, 3, figsize=(8, 8))
vmax = float(xp.max(x))
for i in range(9):
axx = ax5.ravel()[i]
axx.imshow(
parallelproj.to_numpy_array(x[:, i+slice, :]), cmap="Greys", vmin=0, vmax=vmax
)
axx.set_title(f"img plane {i+slice}", fontsize="medium")
# ax5.ravel()[-1].set_axis_off()
fig5.tight_layout()
fig5.show()

























 被折叠的 条评论
为什么被折叠?
被折叠的 条评论
为什么被折叠?








