> library(ggplot2)
> library(tidyverse)
> library(openxlsx)
> library(vegan)
> library(gridExtra)
> df<- read.xlsx('阳加阴种.xlsx',rowNames = T)
> group<-read.xlsx('分组.xlsx',rowNames = T)
> head(df)
T1 T2 T3 T4 T5 T6 T7 T8 T9 N1 N2 N3 N4 N5
[Candida] glabrata 0 0 0 0 0 0 0 25 0 0 0 0 0 0
[Clostridium] innocuum 8 0 62 25 58 3 10 33 36 102 69 29 20 13
[Haemophilus] ducreyi 125 79 109 19 22 93 16 19 46 158 0 115 57 391
[Ruminococcus] gnavus 0 0 218 23 80 4 13 13 52 167 13 79 10 8
Abiotrophia defectiva 59231 34192 191 13472 54 1355 20 268 0 0 0 4601 10372 382
Acholeplasma laidlawii 0 0 0 5 2 0 0 0 0 0 0 0 0 0
> head(group) group T1 P T2 P T3 P T4 P T5 P T6 P
> #计算距离
> distance<-vegdist(df1,method='bray')
> pcoa<- cmdscale(distance,k=(nrow(df1)-1),eig=TRUE)
> plot_data<-data.frame({pcoa$point})[1:3]
> head(plot_data)
X1 X2 X3
T1 -0.24004460 0.002125986 -0.133202721
T2 -0.10770206 -0.016305185 -0.114445307
T3 0.18758419 -0.009082411 -0.122987692
T4 0.16685365 0.062398610 -0.070576883
T5 0.15912776 -0.009955706 -0.110983665
T6 0.04214051 -0.129489736 -0.008899335
> #前三个个分类解释命名
> names(plot_data)[1:3]<-c('PCoA1','PCoA2',"PCoA3")
> eig=pcoa$eig
> group1<-group['group']
> data<-plot_data[match(rownames(group),rownames(plot_data)),]
> data<-data.frame(group,plot_data)
> head(data)
group PCoA1 PCoA2 PCoA3
T1 P -0.24004460 0.002125986 -0.133202721
T2 P -0.10770206 -0.016305185 -0.114445307
T3 P 0.18758419 -0.009082411 -0.122987692
T4 P 0.16685365 0.062398610 -0.070576883
T5 P 0.15912776 -0.009955706 -0.110983665
T6 P 0.04214051 -0.129489736 -0.008899335
> tail(data)
group PCoA1 PCoA2 PCoA3
T9 P 0.12150498 -0.14844320 0.09222156
N1 N -0.11158127 -0.20843923 0.04462288
N2 N 0.13681484 0.15891171 0.01218534
N3 N 0.14604999 -0.01425486 0.19422165
N4 N -0.03413088 0.04983284 0.05242250
N5 N -0.23928394 -0.02998812 0.05641289
|
> data$name = rownames(data)
> t1 <- ggplot(data,aes(x = PCoA1, y = PCoA2, shape = group, color = group))+
+ geom_point()+
+ theme_classic()+
+ geom_vline(xintercept = 0, color = 'gray', size = 0.4) + # 在0处添加垂直线条
+ geom_hline(yintercept = 0, color = 'gray', size = 0.4) +
+ stat_ellipse(aes(x = PCoA1, y = PCoA2 ),
+ geom = "polygon",
+ level = 0.95,
+ alpha=0.4,
+ fill = "transparent")+
+ geom_text( # 添加文本标签
+ aes(label=name),
+ vjust=1.5,
+ size=2,
+ color = "black"
+ )+
+ labs(
+ x = " ",
+ y = paste0("PCoA2 (",as.character(round(pcoa$eig[2] / sum(pcoa$eig) * 100,2)),"%)")
+ )+
+ theme(legend.position = "none")
> t1

> t2 <- ggplot(data,aes(x = PCoA3, y = PCoA2, shape = group, color = group))+
+ geom_point()+
+ theme_classic()+
+ geom_vline(xintercept = 0, color = 'gray', size = 0.4) + # 在0处添加垂直线条
+ geom_hline(yintercept = 0, color = 'gray', size = 0.4) +
+ stat_ellipse(aes(x = PCoA3, y = PCoA2 ),
+ geom = "polygon",
+ level = 0.95,
+ alpha=0.4,
+ fill = "transparent")+
+ geom_text( # 添加文本标签
+ aes(label=name),
+ vjust=1.5,
+ size=2,
+ color = "black"
+ )+
+ labs( # 更改x与y轴坐标为pcoa$eig/sum(pcoa$eig)
+ x = paste0("PCoA3 (",as.character(round(pcoa$eig[3] / sum(pcoa$eig) * 100,2)),"%)"),
+ y = " "
+ )+
+ theme(legend.key.size = unit(0.3, "cm"))
> t2

> t3 <- ggplot(data,aes(x = PCoA1, y = PCoA3, shape = group, color = group))+
+ geom_point()+
+ theme_classic()+
+ geom_vline(xintercept = 0, color = 'gray', size = 0.4) + # 在0处添加垂直线条
+ geom_hline(yintercept = 0, color = 'gray', size = 0.4) +
+ stat_ellipse(aes(x = PCoA1, y = PCoA3 ),
+ geom = "polygon",
+ level = 0.95,
+ alpha=0.4,
+ fill = "transparent")+
+ geom_text( # 添加文本标签
+ aes(label=name),
+ vjust=1.5,
+ size=2,
+ color = "black"
+ )+
+ labs( # 更改x与y轴坐标为pcoa$eig/sum(pcoa$eig)
+ x = paste0("PCoA1 (",as.character(round(pcoa$eig[1] / sum(pcoa$eig) * 100,2)),"%)"),
+ y = paste0("PCoA3 (",as.character(round(pcoa$eig[3] / sum(pcoa$eig) * 100,2)),"%)")
+ )+
+ theme(legend.position = "none")
> t3

> p1_cell <- arrangeGrob(t1)
>
> # 将 p2 放在第二行第一列
> p2_cell <- arrangeGrob(t2)
>
> # 将 p3 放在第二行第二列
> p3_cell <- arrangeGrob(t3)
>
> # 组合图形为四格布局
> plot_grid <- grid.arrange(p1_cell, p2_cell, p3_cell, nrow = 2, ncol = 2,
+ top = " ",
+ widths = unit(c(1, 1), c("null", "null")),
+ heights = unit(c(1, 1), c("null", "null")))
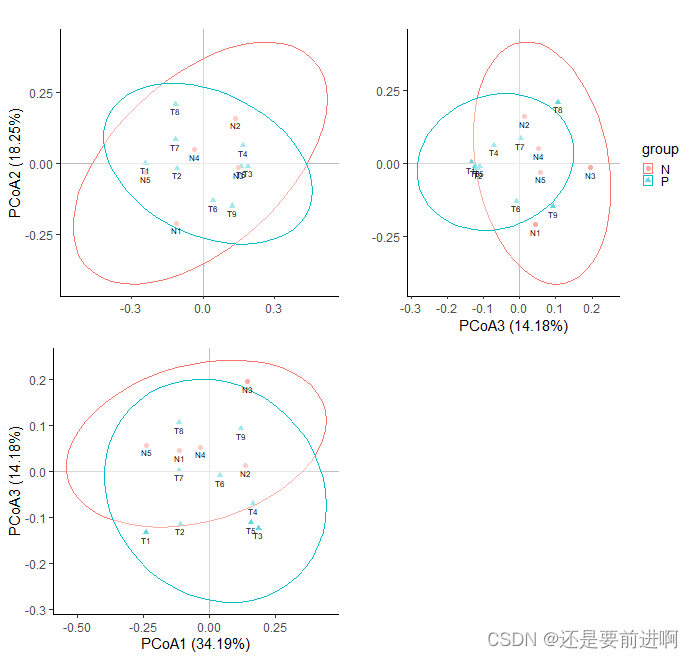
> ggsave("plot.png",plot_grid,dpi = 600, width = 8.0, height = 6.5)

这是将三个PCOA图组合在了一起
























 3115
3115

 被折叠的 条评论
为什么被折叠?
被折叠的 条评论
为什么被折叠?








