K-Means Clustering and Principal Component Analysis
K-Means Clustering总结
- a method to automatically cluster similar data examples together (Unsupervised)
- given a training set { x ( 1 ) , … x ( m ) } \{x^{(1)},\ldots x^{(m)}\} {x(1),…x(m)}, and want to group the data into a few cohesive ‘clusters’.
- iterative procedure that starts by guessing the initial centroids, and then refines this guess by repeatedly assigning examples to their closest centroids and then recomputing the centroids based on the assignments
% Initialize centroids
centroids = kMeansInitCentroids(X, K);
for iter = 1:iterations
% Cluster assignment step: Assign each data point to the
% closest centroid. idx(i) corresponds to c^(i), the index
% of the centroid assigned to example i
idx = findClosestCentroids(X, centroids);
% Move centroid step: Compute means based on centroid
% assignments
centroids = computeMeans(X, idx, K);
end
The inner-loop of the algorithm repeatedly carries out two steps:
- Assigning each training example to its closest centroid
- Recomputing the mean of each centroid using the points assigned to it.
random random initializations
- prevent local optimal
- pick the one yields the lowst distortion (cost function)
% Initialize the centroids to be random examples
%Randomly reorder the indicies of examples
randidx = randperm(size(X,1));
% Take the first K examples
centroids = X(randidx(1:K),:);
Finding closest centroids
c
(
i
)
:
=
j
that minimizes
∣
∣
x
(
i
)
−
μ
j
∣
∣
2
c^{(i)} :=j \quad\text{that minimizes}\quad\left||x^{(i)}-\mu_j\right||^2
c(i):=jthat minimizes∣∣∣x(i)−μj∣∣∣2
MatLab 代码:
function idx = findClosestCentroids(X, centroids)
% Set K (number of centroid)
K = size(centroids, 1);
% number of training examples -- (Mx2) because 2 dimensional,has 2 colums
M = size(X,1);
% You need to return the following variables correctly.
idx = zeros(size(X,1), 1);
for sample = 1:M
distance_array = zeros(1,K); % need a unique distance array for each traning example
% this distance arrau should be a one-dimensional vector (1xK)
for centroid = 1:K
%distance_array(1,centroid) = sqrt (sum(power((X(sample,:)-centroids(centroid,:)),2)));
% another way of calculating this:
distance_array(1, centroid) = ((X(sample,:)' - centroids(centroid,:)').^2)' * ((X(sample,:)' - centroids(centroid,:)').^2);
end
[~, d_idx] = min(distance_array);
idx(sample,1) = d_idx;
end
end
Computing centroid means
function centroids = computeCentroids(X, idx, K)
% Useful variables
[m n] = size(X);
% m-number of training exmaple, n-dimensions
% You need to return the following variables correctly.
centroids = zeros(K, n);
% Instructions: Go over every centroid and compute mean of all points that
% belong to it. Concretely, the row vector centroids(i, :)
% should contain the mean of the data points assigned to
% centroid i.
%idx -- a mx1 vector contains which centroid the example belongs to
for i = 1:K
centroids(i,:) = sum(X((idx==i),:))/length(X((idx==i),:));
end
% method2:
for k=1:K % for-loop over the centroids
centroids(k, :) = mean(X(idx==k, :));
end
end
Principal Component Analysis - Dimensionality reduction
PCA consists of two computational steps:
- compute the covariance(协方差) matrix of the data (denoted as Sigma)
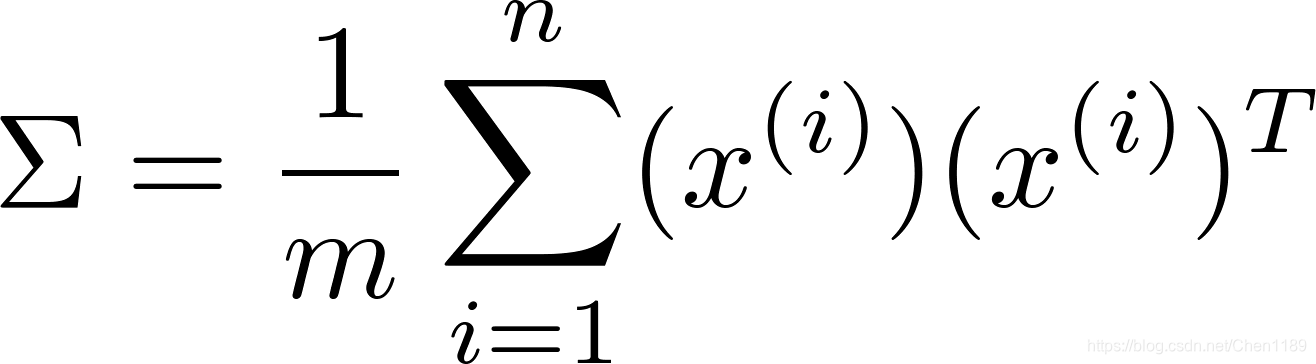
- MATLAB’s svd(Singular value decomposition) function to compute the eigenvectors U 1 , U 2 , … , U n U_1, U_2,\ldots,U_n U1,U2,…,Un --> These will correspond to the principal components of variation in the data.
[U, S, V] = svd(Sigma);
U will contain the principal components and S will contain a diagonal matrix.
Before PCA: preprocessing data:
- normalize the data by subtracting the mean value of each feature from the dataset
- scaling each dimension so that they are in the same range
- feature scaling function in MATLAB (return normalized X and mean value mu, and standard deviation sigma):
function [X_norm, mu, sigma] = featureNormalize(X)
mu = mean(X);
X_norm = bsxfun(@minus, X, mu);
sigma = std(X_norm);
X_norm = bsxfun(@rdivide, X_norm, sigma);
end
PCA implementation
function [U, S] = pca(X)
% Useful values
[m, n] = size(X);
% You need to return the following variables correctly.
U = zeros(n);
S = zeros(n);
% compute covariance matrix (2x300)(300x2) --> 2x2
Sigma = (1.0/m) * X' * X;
[U, S, ~] = svd(Sigma);
end
Dimensionality reduction
project data to lower dimension x ( i ) → z ( i ) x^{(i)}\rightarrow z^{(i)} x(i)→z(i)
Projecting the data onto the principal components
- Given: dataset X, the principal components U, and the desired number of dimensions to reduce to K
- project each example in X onto the top K components in U
- top K components in U are given by the first K columns of U, that is U_reduce = U(:, 1:K)
function Z = projectData(X, U, K)
%PROJECTDATA Computes the reduced data representation when projecting only
%on to the top k eigenvectors
% Z = projectData(X, U, K) computes the projection of
% the normalized inputs X into the reduced dimensional space spanned by
% the first K columns of U. It returns the projected examples in Z.
%
% You need to return the following variables correctly.
Z = zeros(size(X, 1), K);
% ====================== YOUR CODE HERE ======================
% Instructions: Compute the projection of the data using only the top K
% eigenvectors in U (first K columns).
% For the i-th example X(i,:), the projection on to the k-th
% eigenvector is given as follows:
% x = X(i, :)';
% projection_k = x' * U(:, k);
%
% matrix multiplication
Ureduced = U(:, 1:K); % U: nxn, Ureduced: nxK
% the Z we want: mxK, X: mxn
Z = X * Ureduced
% method2:
for i=1:size(X, 1),
for k=1:K,
x = X(i, :)';
projection_k = x' * U(:, k);
Z(i, k) = projection_k;
end
end
end
Recover data
project each example in Z back onto the original space
function X_rec = recoverData(Z, U, K)
% You need to return the following variables correctly.
X_rec = zeros(size(Z, 1), size(U, 1)); % size should be mxn
Ureduced = U (:, 1:K); % nxK matrix
% Z is mxK
X_rec = Z * Ureduced';
% method 2:
for i=1:size(Z, 1),
for j=1:size(U,1),
v = Z(i, :)';
recovered_j = v' * U(j, 1:K)';
X_rec(i, j) = recovered_j;
end
end
end






















 1123
1123

 被折叠的 条评论
为什么被折叠?
被折叠的 条评论
为什么被折叠?








