原理和公式可以去看下面的博客,这里不再赘述。
Lin, et al. Jointneighborhoodentropy-basedgeneselectionmethodwithfisher
scorefortumorclassification. Applied Intelligence.2018.

'''计算Fisher score'''
import numpy as np
from sklearn import datasets
from sklearn.svm import SVC
from sklearn.model_selection import train_test_split
from sklearn.metrics import accuracy_score
class fisherscore():
def __init__(self, X, y):
self.X = X
self.y = y
self.nSample, self.nDim = X.shape
self.labels = np.unique(y)
self.nClass = len(self.labels)
self.total_mean = np.mean(self.X, axis=0)
'''
[mean(a1), mean(a2), ... , mean(am)]
'''
self.class_num, self.class_mean, self.class_std = self.get_mean_std()
'''
std(c1_a1), std(c1_a2), ..., std(c1_am)
std(c2_a1), std(c2_a2), ..., std(c2_am)
std(c3_a1), std(c3_a2), ..., std(c3_am)
'''
self.fisher_score_list = [self.cal_FS(j) for j in range(self.nDim)]
def get_mean_std(self):
Num = np.zeros(self.nClass)
Mean = np.zeros((self.nClass, self.nDim))
Std = np.zeros((self.nClass, self.nDim))
for i, lab in enumerate(self.labels):
idx_list = np.where(self.y == lab)[0]
# print(idx_list[0])
Num[i] = len(idx_list)
Mean[i] = np.mean(self.X[idx_list], axis=0)
Std[i] = np.std(self.X[idx_list], axis=0)
return Num, Mean, Std
def cal_FS(self,j):
Sb_j = 0.0
Sw_j = 0.0
for i in range(self.nClass):
Sb_j += self.class_num[i] * (self.class_mean[i,j] - self.total_mean[j])**2
Sw_j += self.class_num[i] * self.class_std[i,j] **2
return Sb_j / Sw_j
if __name__ == '__main__':
X, y = datasets.load_iris(return_X_y=True)
FS = fisherscore(X, y)
print(FS.class_num)
print(FS.class_mean)
print(FS.class_std)
print(FS.fisher_score_list)
X_train, X_test, y_train, y_test = train_test_split(X,y,test_size=0.3)
model1 = SVC()
model1.fit(X=X_train,y=y_train)
y_hat = model1.predict(X=X_test)
acc = accuracy_score(y_true=y_test,y_pred=y_hat)
print("ACC=",acc)
model1 = SVC()
model1.fit(X=X_train[:,2:],y=y_train)
y_hat = model1.predict(X=X_test[:,2:])
acc = accuracy_score(y_true=y_test,y_pred=y_hat)
print("ACC1=",acc)
model1 = SVC()
model1.fit(X=X_train[:,0:2],y=y_train)
y_hat = model1.predict(X=X_test[:,0:2])
acc = accuracy_score(y_true=y_test,y_pred=y_hat)
print("ACC2=",acc)
结果:
ACC= 0.9555555555555556
ACC1= 0.9555555555555556
ACC2= 0.9111111111111111
再次验证代码的正确性。
X = [[2,2,3,4,5,7],[3,0,1,1,2,4],[4,5,6,9,7,10]]
X = np.array(X)
y = np.array([0,0,1])将例子替换为亨少德小迷弟中的例子。
输出:
[3.0, 5.333333333333334, 5.333333333333334, 6.259259259259259, 1.8148148148148147, 3.0]
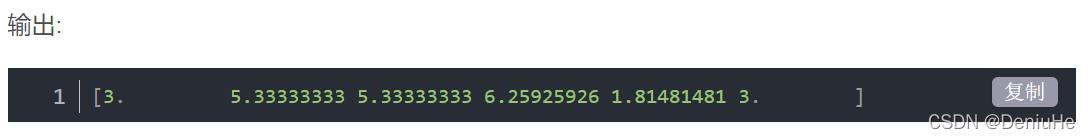
结果一致。
读者可以放心使用。
注: 对数据进行标准化和不做标准化,得到的fisher score 是相同的。
将上述过程写在一个函数中:
import numpy as np
import pandas as pd
class FS():
def __init__(self,X, y, labeled):
self.X = X
self.nDim = X.shape[1]
self.labels = np.unique(y)
self.nClass = len(np.unique(y))
self.y = y
self.labeled = labeled
self.fs = self.attribute_weight()
def attribute_weight(self):
# ------------Initialize some things------------
total_Mean = np.mean(self.X[self.labeled], axis=0)
class_Num = np.zeros(self.nClass)
class_Mean = np.zeros((self.nClass, self.nDim))
class_std = np.zeros((self.nClass, self.nDim))
fisher_score_list = np.zeros(self.nDim)
# --------calculate intermediate components---------
labeled_y = self.y[self.labeled]
labeled_idx = np.array(self.labeled)
for i, lab in enumerate(self.labels):
ids = np.where(labeled_y == lab)[0]
lab_ids = labeled_idx[ids] # 类别为lab的已标记样本索引
class_Num[i] = len(lab_ids)
class_Mean[i] = np.mean(self.X[lab_ids], axis=0)
class_std[i] = np.std(self.X[lab_ids], axis=0)
# --utlize the intermediate components to compute the FS------
for j in range(self.nDim):
Sb_j = 0.0
Sw_j = 0.0
for i in range(self.nClass):
Sb_j += class_Num[i] * (class_Mean[i,j] - total_Mean[j]) ** 2
Sw_j += class_Num[i] * class_std[i,j] ** 2
fisher_score_list[j] = Sb_j / Sw_j
print("FS::",fisher_score_list)
# -----softmax based normalize FS range to [0,1]------
Norm_fs = np.exp(fisher_score_list)
Norm_fs_sum = sum(Norm_fs)
Norm_fs = Norm_fs / Norm_fs_sum
return Norm_fs
X = [[2,2,3,4,5,7],[3,0,1,1,2,4],[4,5,6,9,7,10]]
X = np.array(X)
y = np.array([0,0,1])
model = FS(X=X,y=y, labeled=[0,1,2])
print(model.fs)
print(sum(model.fs))







 https://blog.csdn.net/qq_39923466/article/details/118809782
https://blog.csdn.net/qq_39923466/article/details/118809782
















 4171
4171

 被折叠的 条评论
为什么被折叠?
被折叠的 条评论
为什么被折叠?










