探索过程:
了解fingerprint是什么:
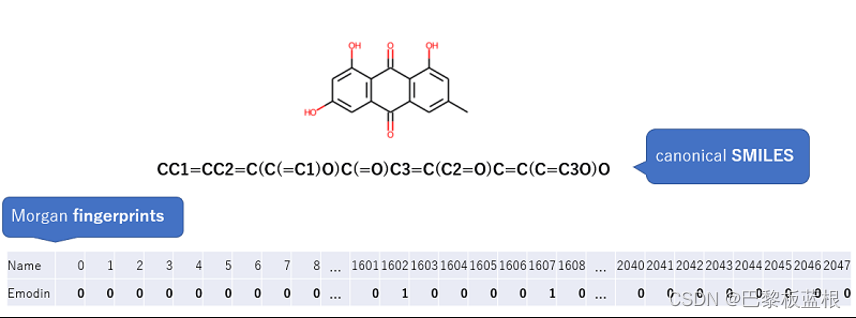
输入:只包含一列的SMILES文件,命名为‘SMILES.txt’(列名为SMILES)
以下是test,均未成功:
我苦苦寻找 rdkit的chem模块教程,转出来结果一脸懵逼。
test1: 引用自这里https://cloud.tencent.com/developer/ask/sof/342250,代码如下:
from rdkit import Chem
import pandas as pd
import rdkit
from rdkit.Chem import AllChem
from rdkit import Chem,DataStructs
import seaborn as sns
from scipy import stats
import matplotlib.pyplot as plt
data = pd.read_csv('SMILE.txt')
smiles_list = data.SMILES.tolist()
# create a list of mols
mols = [Chem.MolFromSmiles(smiles) for smiles in smiles_list]
# create a list of fingerprints from mols
fps = [Chem.RDKFingerprint(mol) for mol in mols]
print(fps)
test2: 引用自这里https://blog.csdn.net/gongfuxiongmao_/article/details/125336253,代码如下:
# -* - coding: UTF-8 -* -
import pandas as pd
import rdkit
from rdkit.Chem import AllChem
from rdkit import Chem,DataStructs
import seaborn as sns
from scipy import stats
import matplotlib.pyplot as plt
data = pd.read_csv('SMILE.txt')
data_1 = data.SMILES.tolist()
# 生成摩根指纹函数
def product_fps(data):
"""传入smiles编码文件列表"""
data = [x for x in data if x is not None]
data_mols = [Chem.MolFromSmiles(s) for s in data]
data_mols = [x for x in data_mols if x is not None]
data_fps = [AllChem.GetMorganFingerprintAsBitVect(x,3,2048) for x in data_mols]
return data_fps
# 计算分子相似度函数
def similar(data):
"""传入smiles编码文件列表"""
fps = product_fps(data)
similarity = []
for i in range(len(fps)):
sims = DataStructs.BulkTanimotoSimilarity(fps[i],fps[:i])
similarity.extend(sims)
return similarity
# 函数调用并打印
fps = product_fps(data_1)
# fps.to_csv('./fingerprint_output.csv',index=False,header=False)
print(f'fps:{fps[:20]}')
x=open('./fingerprint_output.txt','w')
x.write(str(fps))
x.close()
similarity = similar(data_1)
# similarity.to_csv('./similarity_output.csv',index=False,header=False)
结果长这个样子:
!!此时发现一个无人问津的教程,耐下性子使劲看从未了解过的python代码,因已无路可退,最终成功,引用自这里https://www.jianshu.com/p/3107a7f4ab78
各分子描述符+RDKit | 基于分子指纹的分子相似性 https://zhuanlan.zhihu.com/p/86701228
Test1:转MASCC
MACCS Keys,由MDL开发的化学结构数据库衍生的指纹,以化学信息学闻名。
1.首先保存这个文件,命名为‘MACCS.py’
import pandas as pd
from rdkit import Chem
from rdkit.Chem import MACCSkeys
class MACCS:
def __init__(self, smiles):
self.smiles = smiles
self.mols = [Chem.MolFromSmiles(i) for i in smiles]
def compute_MACCS(self, name):
MACCS_list = []
header = ['bit' + str(i) for i in range(167)]
for i in range(len(self.mols)):
ds = list(MACCSkeys.GenMACCSKeys(self.mols[i]).ToBitString())
MACCS_list.append(ds)
df = pd.DataFrame(MACCS_list,columns=header)
df.to_csv(name[:-4]+'_MACCS.csv', index=False)
2.然后运行以下代码(注意输入文件的要求格式):
import pandas as pd
from molvs import standardize_smiles
from MACCS import *
def main():
filename = './SMILE.txt' # path to your csv file
df = pd.read_csv(filename) # read the csv file as pandas data frame
smiles = [standardize_smiles(i) for i in df['SMILES'].values]
## Compute MACCS Fingerprints and export file.
maccs_descriptor = MACCS(smiles) # create your MACCS object and provide smiles
maccs_descriptor.compute_MACCS(filename) # 计算 MACCS 并且给文件命名。 命名可以直接使用初始名称,因为MACCS的类会在其后添加 "_MACCS.csv" 作为输出文件。
if __name__ == '__main__':
main()
就得到这个样子的结果了:
共检查了166个子结构,由于1位用于保存RDKit中的信息,因此指纹总共为167位。如果具有子结构,则存储1,否则存储0。
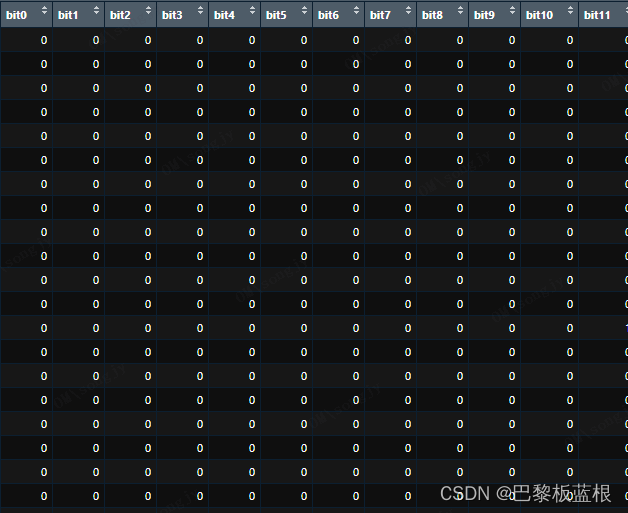
嗯!看起来是我想要的
具体每一列是什么,看这里https://github.com/openbabel/openbabel/blob/master/data/MACCS.txt
虽然都看不懂,但我用起来了。先存一下慢慢看:
Rdkit基础操作 https://blog.csdn.net/qq_40943760/article/details/120345448
RDKit|分子指纹提取、相似性比较及应用:这篇好像简单明白些 https://www.jianshu.com/p/b0148c74e85d
Test2:转ECFP6
1.首先保存这个文件,命名为‘ECFP6.py’
import numpy as np
import pandas as pd
from rdkit.Chem import AllChem
from rdkit import Chem, DataStructs
class ECFP6:
def __init__(self, smiles):
self.mols = [Chem.MolFromSmiles(i) for i in smiles]
self.smiles = smiles
def mol2fp(self, mol, radius = 3):
fp = AllChem.GetMorganFingerprintAsBitVect(mol, radius = radius)
array = np.zeros((1,))
DataStructs.ConvertToNumpyArray(fp, array)
return array
def compute_ECFP6(self, name):
bit_headers = ['bit' + str(i) for i in range(2048)]
arr = np.empty((0,2048), int).astype(int)
for i in self.mols:
fp = self.mol2fp(i)
arr = np.vstack((arr, fp))
df_ecfp6 = pd.DataFrame(np.asarray(arr).astype(int),columns=bit_headers)
df_ecfp6.insert(loc=0, column='smiles', value=self.smiles)
df_ecfp6.to_csv(name[:-4]+'_ECFP6.csv', index=False)
2.然后运行以下代码(注意输入文件的要求格式):
import pandas as pd
from molvs import standardize_smiles
from ECFP6 import *
def main():
filename = './SMILE.txt' # path to your csv file
df = pd.read_csv(filename) # read the csv file as pandas data frame
smiles = [standardize_smiles(i) for i in df['SMILES'].values]
## Compute ECFP6 Fingerprints and export a csv file.
ecfp6_descriptor = ECFP6(smiles) # create your ECFP6 object and provide smiles
ecfp6_descriptor.compute_ECFP6(filename) # compute ECFP6 and provide the name of your desired output file. you can use the same name as the input file because the ECFP6 class will ensure to add "_ECFP6.csv" as part of the output file.
if __name__ == '__main__':
main()
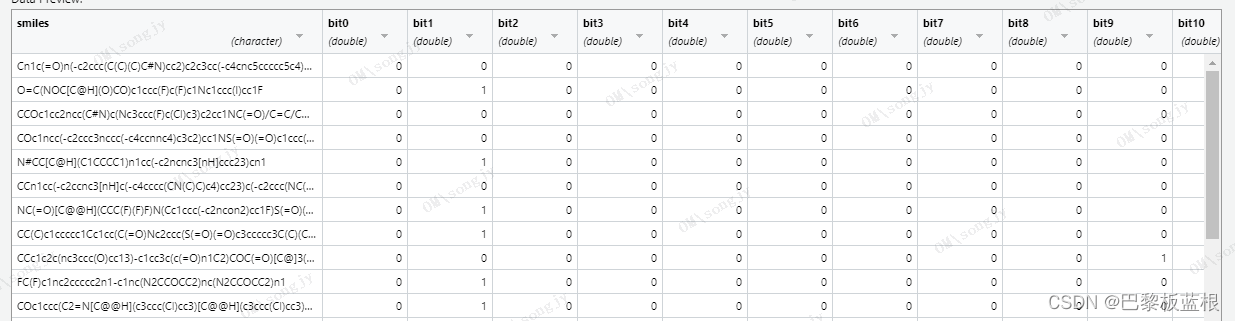
结果是这个样子的,感觉还行






















 38
38











 被折叠的 条评论
为什么被折叠?
被折叠的 条评论
为什么被折叠?








