#PCA的衡量指标是样本方差
#为什么样本方差的分母是n-1?
#简述:因为均值已经用了n个数的平均来做估计在求方差时,只有(n-1)个数和均值信息是不相关的。
#而第n个数已经可以由前(n-1)个数和均值 来唯一确定,实际上没有信息量。所以在计算方差时,只除以(n-1)。
#如何“正确”的估计方差(使得样本方差接近实际总体数据的方差)
#(答案是把样本差值平方之和/n)的分母n换成n-1,通过这种方法把原来的偏小的估计“放大”一点点,我们就能获得对方差的正确估计了:
#pca与特征选择的区别,新特征矩阵不具有可读性
#2.2重要参数n_components(成分)
#2.2.1高维数据可视化
import matplotlib.pyplot as plt
from sklearn.datasets import load_iris
from sklearn.decomposition import PCA #降维模块
iris=load_iris()
y=iris.target
X=iris.data
X.shape
(150, 4)
import pandas as pd
pd.DataFrame(X)
| 0 | 1 | 2 | 3 | |
|---|---|---|---|---|
| 0 | 5.1 | 3.5 | 1.4 | 0.2 |
| 1 | 4.9 | 3.0 | 1.4 | 0.2 |
| 2 | 4.7 | 3.2 | 1.3 | 0.2 |
| 3 | 4.6 | 3.1 | 1.5 | 0.2 |
| 4 | 5.0 | 3.6 | 1.4 | 0.2 |
| ... | ... | ... | ... | ... |
| 145 | 6.7 | 3.0 | 5.2 | 2.3 |
| 146 | 6.3 | 2.5 | 5.0 | 1.9 |
| 147 | 6.5 | 3.0 | 5.2 | 2.0 |
| 148 | 6.2 | 3.4 | 5.4 | 2.3 |
| 149 | 5.9 | 3.0 | 5.1 | 1.8 |
150 rows × 4 columns
y
array([0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1,
1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1,
1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2,
2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2,
2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2])
pca=PCA(n_components=2)
X_dr=pca.fit_transform(X)
X_dr
array([[-2.68412563, 0.31939725],
[-2.71414169, -0.17700123],
[-2.88899057, -0.14494943],
[-2.74534286, -0.31829898],
[-2.72871654, 0.32675451],
[-2.28085963, 0.74133045],
[-2.82053775, -0.08946138],
[-2.62614497, 0.16338496],
[-2.88638273, -0.57831175],
[-2.6727558 , -0.11377425],
[-2.50694709, 0.6450689 ],
[-2.61275523, 0.01472994],
[-2.78610927, -0.235112 ],
[-3.22380374, -0.51139459],
[-2.64475039, 1.17876464],
[-2.38603903, 1.33806233],
[-2.62352788, 0.81067951],
[-2.64829671, 0.31184914],
[-2.19982032, 0.87283904],
[-2.5879864 , 0.51356031],
[-2.31025622, 0.39134594],
[-2.54370523, 0.43299606],
[-3.21593942, 0.13346807],
[-2.30273318, 0.09870885],
[-2.35575405, -0.03728186],
[-2.50666891, -0.14601688],
[-2.46882007, 0.13095149],
[-2.56231991, 0.36771886],
[-2.63953472, 0.31203998],
[-2.63198939, -0.19696122],
[-2.58739848, -0.20431849],
[-2.4099325 , 0.41092426],
[-2.64886233, 0.81336382],
[-2.59873675, 1.09314576],
[-2.63692688, -0.12132235],
[-2.86624165, 0.06936447],
[-2.62523805, 0.59937002],
[-2.80068412, 0.26864374],
[-2.98050204, -0.48795834],
[-2.59000631, 0.22904384],
[-2.77010243, 0.26352753],
[-2.84936871, -0.94096057],
[-2.99740655, -0.34192606],
[-2.40561449, 0.18887143],
[-2.20948924, 0.43666314],
[-2.71445143, -0.2502082 ],
[-2.53814826, 0.50377114],
[-2.83946217, -0.22794557],
[-2.54308575, 0.57941002],
[-2.70335978, 0.10770608],
[ 1.28482569, 0.68516047],
[ 0.93248853, 0.31833364],
[ 1.46430232, 0.50426282],
[ 0.18331772, -0.82795901],
[ 1.08810326, 0.07459068],
[ 0.64166908, -0.41824687],
[ 1.09506066, 0.28346827],
[-0.74912267, -1.00489096],
[ 1.04413183, 0.2283619 ],
[-0.0087454 , -0.72308191],
[-0.50784088, -1.26597119],
[ 0.51169856, -0.10398124],
[ 0.26497651, -0.55003646],
[ 0.98493451, -0.12481785],
[-0.17392537, -0.25485421],
[ 0.92786078, 0.46717949],
[ 0.66028376, -0.35296967],
[ 0.23610499, -0.33361077],
[ 0.94473373, -0.54314555],
[ 0.04522698, -0.58383438],
[ 1.11628318, -0.08461685],
[ 0.35788842, -0.06892503],
[ 1.29818388, -0.32778731],
[ 0.92172892, -0.18273779],
[ 0.71485333, 0.14905594],
[ 0.90017437, 0.32850447],
[ 1.33202444, 0.24444088],
[ 1.55780216, 0.26749545],
[ 0.81329065, -0.1633503 ],
[-0.30558378, -0.36826219],
[-0.06812649, -0.70517213],
[-0.18962247, -0.68028676],
[ 0.13642871, -0.31403244],
[ 1.38002644, -0.42095429],
[ 0.58800644, -0.48428742],
[ 0.80685831, 0.19418231],
[ 1.22069088, 0.40761959],
[ 0.81509524, -0.37203706],
[ 0.24595768, -0.2685244 ],
[ 0.16641322, -0.68192672],
[ 0.46480029, -0.67071154],
[ 0.8908152 , -0.03446444],
[ 0.23054802, -0.40438585],
[-0.70453176, -1.01224823],
[ 0.35698149, -0.50491009],
[ 0.33193448, -0.21265468],
[ 0.37621565, -0.29321893],
[ 0.64257601, 0.01773819],
[-0.90646986, -0.75609337],
[ 0.29900084, -0.34889781],
[ 2.53119273, -0.00984911],
[ 1.41523588, -0.57491635],
[ 2.61667602, 0.34390315],
[ 1.97153105, -0.1797279 ],
[ 2.35000592, -0.04026095],
[ 3.39703874, 0.55083667],
[ 0.52123224, -1.19275873],
[ 2.93258707, 0.3555 ],
[ 2.32122882, -0.2438315 ],
[ 2.91675097, 0.78279195],
[ 1.66177415, 0.24222841],
[ 1.80340195, -0.21563762],
[ 2.1655918 , 0.21627559],
[ 1.34616358, -0.77681835],
[ 1.58592822, -0.53964071],
[ 1.90445637, 0.11925069],
[ 1.94968906, 0.04194326],
[ 3.48705536, 1.17573933],
[ 3.79564542, 0.25732297],
[ 1.30079171, -0.76114964],
[ 2.42781791, 0.37819601],
[ 1.19900111, -0.60609153],
[ 3.49992004, 0.4606741 ],
[ 1.38876613, -0.20439933],
[ 2.2754305 , 0.33499061],
[ 2.61409047, 0.56090136],
[ 1.25850816, -0.17970479],
[ 1.29113206, -0.11666865],
[ 2.12360872, -0.20972948],
[ 2.38800302, 0.4646398 ],
[ 2.84167278, 0.37526917],
[ 3.23067366, 1.37416509],
[ 2.15943764, -0.21727758],
[ 1.44416124, -0.14341341],
[ 1.78129481, -0.49990168],
[ 3.07649993, 0.68808568],
[ 2.14424331, 0.1400642 ],
[ 1.90509815, 0.04930053],
[ 1.16932634, -0.16499026],
[ 2.10761114, 0.37228787],
[ 2.31415471, 0.18365128],
[ 1.9222678 , 0.40920347],
[ 1.41523588, -0.57491635],
[ 2.56301338, 0.2778626 ],
[ 2.41874618, 0.3047982 ],
[ 1.94410979, 0.1875323 ],
[ 1.52716661, -0.37531698],
[ 1.76434572, 0.07885885],
[ 1.90094161, 0.11662796],
[ 1.39018886, -0.28266094]])
X_dr.shape
(150, 2)
y==0
array([ True, True, True, True, True, True, True, True, True,
True, True, True, True, True, True, True, True, True,
True, True, True, True, True, True, True, True, True,
True, True, True, True, True, True, True, True, True,
True, True, True, True, True, True, True, True, True,
True, True, True, True, True, False, False, False, False,
False, False, False, False, False, False, False, False, False,
False, False, False, False, False, False, False, False, False,
False, False, False, False, False, False, False, False, False,
False, False, False, False, False, False, False, False, False,
False, False, False, False, False, False, False, False, False,
False, False, False, False, False, False, False, False, False,
False, False, False, False, False, False, False, False, False,
False, False, False, False, False, False, False, False, False,
False, False, False, False, False, False, False, False, False,
False, False, False, False, False, False, False, False, False,
False, False, False, False, False, False])
colors=['red','black','orange']
plt.figure()
for i in[0,1,2]:
plt.scatter(X_dr[y==i,0]
,X_dr[y==i,1]
,alpha=.7
,c=colors[i]
,label=iris.target_names[i]
)
plt.legend()#画图例
plt.title('PCA of IRIS dataset')
plt.show()
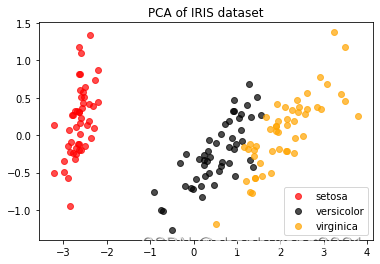
pca.explained_variance_#(信息量,可解释性方差)
array([4.22824171, 0.24267075])
pca.explained_variance_ratio_#特征在原数据的信息占比
array([0.92461872, 0.05306648])
pca.explained_variance_ratio_.sum()#特征在原数据的信息占比总和
0.9776852063187949
pca_line=PCA().fit(X)
pca_line.explained_variance_ratio_#查看原数据特征信息占比
array([0.92461872, 0.05306648, 0.01710261, 0.00521218])
import numpy as np
np.cumsum(pca_line.explained_variance_ratio_)
array([0.92461872, 0.97768521, 0.99478782, 1. ])
pac_line=PCA().fit(X)
plt.plot([1,2,3,4],np.cumsum(pca_line.explained_variance_ratio_))
plt.xticks([1,2,3,4])#使得坐标轴显示为整数
plt.xlabel("number of components after dimension reduction")
plt.ylabel("cumulative explained variance ratip")
plt.show()
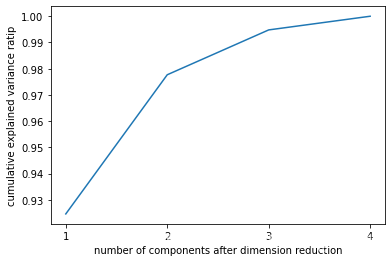
pca_mle=PCA(n_components='mle')
pca_mle=pca_mle.fit(X)
X_mle=pca_mle.transform(X)
X_mle.shape
(150, 3)
pca_mle.explained_variance_ratio_.sum()
0.9947878161267246
#2.2.3 按信息占比选超参数
pca_f=PCA(n_components=0.97,svd_solver='full')
pca_f=pca_f.fit(X)
X_f=pca_f.transform(X)
pca_f.explained_variance_ratio_
array([0.92461872, 0.05306648])
pca_f.explained_variance_ratio_.sum()
0.9776852063187949
#2.3PCA中的SVD
#2.3.1PCA中的SVD哪里来?
#奇异值分解跳过计算协方差矩阵直接求出新空间(空间转换矩阵)和降维后的特征矩阵
#在矩阵分解时不使用PCA本身的特征值分解,而使用奇异值分解来减少计算量,奇异值分解产生的V(k,n)保存于属性componenets_
PCA(2).fit(X).components_
array([[ 0.36138659, -0.08452251, 0.85667061, 0.3582892 ],
[ 0.65658877, 0.73016143, -0.17337266, -0.07548102]])
PCA(2).fit(X).components_.shape#降维后的新特征空间
(2, 4)
#2.3.2svd_solver与random_state
#full:根据原始数据和n_components计算寻找特征向量,完整的生成SVD的三个结构
#auto:如果数据大于500*500且特征数小于数据最小维度80%,就启用randomized截断会在矩阵被分解后有选择的发生
#arpack:适用于高维的稀疏矩阵,加快分解速度
#randomized:适用于巨大矩阵,计算量大;分解器生成随机向量,检测是否符合需求,如果符合,保留随机向量,基于此构建后续向量空间
#random_state在参数svd_solver为'arpack'or'randomized'
#如果特征空间是图像,空间矩阵能够可视化
from sklearn.datasets import fetch_lfw_people
from sklearn.decomposition import PCA
import matplotlib.pyplot as plt
import numpy as np
faces=fetch_lfw_people(min_faces_per_person=60)#实例化
faces.data.shape#降维支持小于二维特征,(images形式)
(1348, 2914)
faces.images.shape#1348是图像个数,62,47分别是特征矩阵的行列
(1348, 62, 47)
X=faces.data
fig,axes=plt.subplots(4,5
,figsize=(8,4)
,subplot_kw={"xticks":[],"yticks":[]}
)
axes[0][0].imshow(faces.images[0,:,:])#轴图像(子画布)
<matplotlib.image.AxesImage at 0x1d6b4480c70>
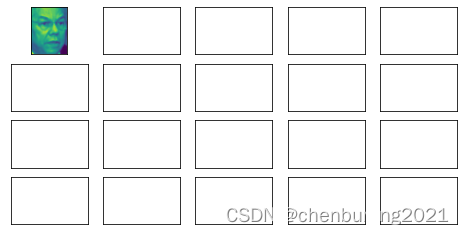
axes.shape
(4, 5)
axes[0][0].imshow(faces.images[0,:,:])
<matplotlib.image.AxesImage at 0x1d6b44803a0>
axes.flat
<numpy.flatiter at 0x1d6add0a890>
([*axes.flat])
[<AxesSubplot:>,
<AxesSubplot:>,
<AxesSubplot:>,
<AxesSubplot:>,
<AxesSubplot:>,
<AxesSubplot:>,
<AxesSubplot:>,
<AxesSubplot:>,
<AxesSubplot:>,
<AxesSubplot:>,
<AxesSubplot:>,
<AxesSubplot:>,
<AxesSubplot:>,
<AxesSubplot:>,
<AxesSubplot:>,
<AxesSubplot:>,
<AxesSubplot:>,
<AxesSubplot:>,
<AxesSubplot:>,
<AxesSubplot:>]
len([*axes.flat])#[*]打开惰性对象,.flat降维
20
#填充函数
[*enumerate(axes.flat)]#enumerate形成元组
[(0, <AxesSubplot:>),
(1, <AxesSubplot:>),
(2, <AxesSubplot:>),
(3, <AxesSubplot:>),
(4, <AxesSubplot:>),
(5, <AxesSubplot:>),
(6, <AxesSubplot:>),
(7, <AxesSubplot:>),
(8, <AxesSubplot:>),
(9, <AxesSubplot:>),
(10, <AxesSubplot:>),
(11, <AxesSubplot:>),
(12, <AxesSubplot:>),
(13, <AxesSubplot:>),
(14, <AxesSubplot:>),
(15, <AxesSubplot:>),
(16, <AxesSubplot:>),
(17, <AxesSubplot:>),
(18, <AxesSubplot:>),
(19, <AxesSubplot:>)]
fig,axes=plt.subplots(3,8
,figsize=(8,4)
,subplot_kw={"xticks":[],"yticks":[]}
)
for i,ax in enumerate(axes.flat):
ax.imshow(faces.images[i,:,:]
,cmap="gray")

#4.降维,提取特征空间
pca=PCA(150).fit(X)
V=pca.components_#V(k,n)保存在属性components_中:新特征矩阵,转化矩阵,可以将原特征矩阵转化新矩阵
V.shape
(150, 2914)
fig,axes=plt.subplots(3,8
,figsize=(8,4)
,subplot_kw={"xticks":[],"yticks":[]}
)
for i,ax in enumerate(axes.flat):
ax.imshow(V[i,:].reshape(62,47)#一维特征升二维特征,形式改变可视化
,cmap="gray")
#显示了新特征空间的特点,新特征为重要数据[五官,明暗分布]

pca=PCA(150)
X_dr=pca.fit_transform(X)
X_dr.shape
#获得新特征
(1348, 150)
X_inverse=pca.inverse_transform(X_dr )
X_inverse.shape
#将新特征导回原空间,升维,图像于原图像高度相似,但是信息仍损失,图片变模糊,所以降维是不可逆的
(1348, 2914)
fig,ax=plt.subplots(2,10,figsize=(10,2.5)
,subplot_kw={"xticks":[],"yticks":[]}
)
for i in range(10):
ax[0,i].imshow(faces.images[i,:,:],cmap="binary_r")
ax[1,i].imshow(X_inverse[i].reshape(62,47),cmap="binary_r")

ax[0,1]
<AxesSubplot:>
faces.images.shape
(1348, 62, 47)
#2.4.2PCA做噪音过滤
from sklearn.datasets import load_digits
from sklearn.decomposition import PCA
import matplotlib.pyplot as plt
import numpy as np
digits=load_digits()
digits.data.shape
(1797, 64)
set(digits.target.tolist())#去重
{0, 1, 2, 3, 4, 5, 6, 7, 8, 9}
digits.images.shape
(1797, 8, 8)
def plot_digits(data):#设置封装接口
fig,axes=plt.subplots(4,10,figsize=(10,4)
,subplot_kw={"xticks":[],"yticks":[]}
)
for i,ax in enumerate(axes.flat):
ax.imshow(data[i].reshape(8,8),cmap="binary")
plot_digits(digits.data)

import numpy as np
rng=np.random.RandomState(42) #规定随机模式
noisy=rng.normal(digits.data,2)#从中随机抽取满足正态分布的另一个数据集,剥离创造,和设置抽取正态分布的标准差
noisy.shape#(同原数据结构)
(1797, 64)
plot_digits(noisy)

pca=PCA(0.5,svd_solver="full").fit(noisy)#获得大量有效特征
X_dr=pca.transform(noisy)
X_dr.shape
(1797, 6)
without_noise=pca.inverse_transform(X_dr)
without_noise.shape
(1797, 64)
plot_digits(without_noise)

#重要参数
#n_components:成分,svd_solver:svd处理方法,randomized控制拥有随机模型的处理方法
#三个重要属性
#components:V(k,n)保存在属性components_中:转化矩阵;
#explained_variance:解释性特征
#explained_variance_ratio:解释性特征信息占比
#接口inverse_transform,降维恢复(信息仍损失)#将降维后数据返回原特征空间
#没啥用,计算协方差矩阵并计算get_covariance






















 5156
5156











 被折叠的 条评论
为什么被折叠?
被折叠的 条评论
为什么被折叠?








