
提示:文章写完后,目录可以自动生成,如何生成可参考右边的帮助文档
前言
遗传算法作业,弄了一周多,先是跟着羽丶千落大佬的进化算法(GA)Python实现–以ackley函数为例的文章跟了一遍,理解ga的步骤与思路,之后再按老师要求进行了编码。期间遇到的无数困难,为免之后学弟学妹查找资料学习麻烦,也为了未来可能忘了ga怎么弄的我,故而写此博客,毕竟好记性不如烂笔头嘛。
PS.Python老师教的极快,所以实现完ga后,我仍感觉自己很菜,对numpy,matplotlib等库都只是初步了解。所以在第二节我的注释在专业大佬面前看起来可能就是脱裤子放屁,还请理解。
PPS.如果你有任何问题欢迎评论区留言。还有如果你觉得我这篇博客有帮助的话能否请你在评论区说明一下哪里帮到你了呢?或者说你觉得我哪里能改得更好,都可以。非常感谢。
提示:以下是本篇文章正文内容,下面案例可供参考
一、GA算法是什么?
GA算法(Genetic Algorithm 遗传算法)是通过模仿生物繁衍、DNA的编译和自然选择而形成的一种进化算法。是的,进化算法,这个进化指的就是朝着更好(更适应环境)的结果发展。本质上可以理解为一种通过干预来求最优解的算法。
其关键步骤有:
1.初始化种群
即设置一个种群的大小(pop_size),个体维度(dim),当然我本人将维度理解为个体的特征数量,特征的取值范围(bounds)
2.计算种群函数值
通过将种群的每一个个体传入要求解的函数中算出每个个体的函数值。
3. 计算种群适应值
一般通过对种群函数值进行归一化处理(这个连接里有解释什么是归一化和为什么要归一化)来计算种群适应值,当然,直接以种群函数值来做适应值也可以。不过一般会归一化。
4.选择父代个体(也可以理解为自然选择)
被选中的个体才有资格繁衍,可以理解为没被选中的死了。
5.杂交操作
被选中的个体进行繁衍,其基因型进行杂交。比如:A个体基因为00001111,B个体基因为1111000,杂交位置在第4和5间位置进行。那么杂交后,其产生的后代C、D基因型为:11111111,00000000。
6.变异操作
个体的基因进行变异,比如A个体基因11101111,如果其在pos=3(从0开始数)的位置变异的话,其变异后的基因型为11111111。
7.更新种群
如题,将被选中的父代和产生的子代合并为一个种群(个体数量不变,所以要注意控制杂交操作时产生的子代数量)
8.获取最佳个体
将新种群中的适应值最高的个体选出来。
二、羽、千落大佬代码的分析(若大佬觉得不合适,我会把第二节删掉)
基本上就是把大佬的代码进行了比较详细的注释
1.初始化种群
'''
初始化种群大小
种群大小popsize:设置种群大小
维度dim:个体的维度
取值范围bounds:每个个体每一维的取值范围
'''
# PS大佬这里的函数名叫initPop,其他未改
def create_pop(pop_size,dim,bounds):
pop = np.random.rand(pop_size,dim) #生成一个随机的pop_size行dim列矩阵
for i in range(0,dim):
pop[:,i] = pop[:,i]*(bounds[i,1]-bounds[i,0])+bounds[i,0] #对每一行进行赋值
#可改,每行每列乘个数罢了
return pop
2.计算种群函数值
'''
种群计算,计算ackley函数,可以改为其他,ackley函数改一下即可
种群pop:传入种群矩阵(数组)
种群函数值pop_value:返回计算结果
'''
def calAckLey(pop):
pop_x = pop.shape[0] #获取pop的行数,即pop_size
pop_value = np.zeros((pop_x,1),np.float64) #生成全为0的矩阵,之所以只有一列,因为是求y值,只需一个
for i in range(0,pop_x):
pop_value[i,0] = ackley(pop[i,:],dim) #对每一列求值然后给到,这里的pop[i,:]是一行单独作为一个数组传入ackley函数计算
return pop_value
3.计算种群适应值
# 计算种群适应值,值进行归一化处理,,有些父代选择方式需要先做归一化处理
def calFitvalue(pop_value):
value = 1/(pop_value + 0.0001) #防止分母为0
max_value = max(value)
min_value = min(value)
fitvalue = (value - min_value)/(max_value - min_value + 0.001) #防止最大值最小值相同导致为0,ps:这个fit数组里必有一个数是0
return fitvalue
4.选择父代个体(自然选择)
这里决定了你选的是最高值还是最低值,具体在代码中的PS的地方
def selectParent(pop,fitvalue):
fit_index = np.argsort(fitvalue, axis =0) #按行从小到大排列,返回对应元素排之前的下标
fit_index = fit_index[::-1] #此时是从大到小的坐标了 PS,这里通过对fit值方向的排序决定你选的是最大值还是最小值
pop_x,pop_y = pop.shape #pop_x获取行数,pop_y获取列数,或许写成 pop_x = pop.shape[0] pop_y = pop.shape[1]更直观一点?
parent_num = int(pop_x//1.2) #//表示向下取整的最大数 ,不是所有人都可以交配的拜托
#ps我觉得这里写成parent_num = int(pop_x * 0.8)更好更准确,pps选多少作为父代是自定义的
new_pop = np.zeros((parent_num,pop_y),np.float64) #列数不变,但新的pop行数小了,也就是个体少了
for i in range(0,parent_num):
new_pop[i,:] = pop[fit_index[i],:] #直接让子代成为父母中最优的那几个
return new_pop
5.杂交操作
这里大佬用的是实数杂交,所以大佬并未提到进制转换的问题(我后面用的是二进制杂交)。
def crossover(pop,cross_rate):
pop_x,pop_y = pop.shape
child_pop = np.zeros((2,pop_y),dtype = np.float64)
new_pop = np.zeros((1,pop_y),dtype = np.float64)
for i in range(0,pop_x):
if np.random.rand() < cross_rate : #落在cross_rate范围内就交配了
parent_pop1 = pop[i,:]
parent_pop2 = pop[np.random.randint(pop_x),:] # 有可能选到自己哦
n = np.random.rand() #随机截取父母长度
child_pop[0,:] = parent_pop1*n + parent_pop2*(1-n)
child_pop[1,:] = parent_pop1*(1-n) + parent_pop2*n
new_pop = np.concatenate((new_pop,child_pop),axis = 0) #其实直接输出就行,但返回的是new_pop所以得删掉首行的0
new_pop = np.delete(new_pop,0,axis = 0) #删掉第一行全0行
return new_pop
6.变异操作
def mutation(pop,mutation_rate,bounds):#注意,pop或者mutation_rate要设好,自己测试的时候可以设大一点,不然样本少了就啥都没
pop_x,pop_y = pop.shape
new_pop = np.zeros((1,pop_y),dtype = np.float64)
for i in range(0,pop_x):
if np.random.rand() < mutation_rate:
new_pop_m = pop[i,:] #取出第i行
for j in range(0,pop_y):
if np.random.rand() < mutation_rate:
new_pop_m[j] = np.random.rand()*(bounds[j,1] - bounds[j,0]) + bounds[j,0] #第i行j个元素变异,方法和create_pop的方法一样
new_pop = np.append(new_pop,[new_pop_m],axis = 0)
new_pop = np.delete(new_pop,0,axis = 0)
return new_pop
7.种群更新操作
def updatePop(pop,pop_size,fitvalue):
pop_x,pop_y = pop.shape
if pop_x < pop_size: #比之前的小,代表没把原来的pop和变异pop合起来
return pop
new_pop = np.zeros((pop_size,pop_y),dtype = np.float64)
new_fitvalue = np.zeros((pop_size,1),dtype = np.float64)
fit_index = np.argsort(fitvalue,axis = 0)
fit_index = fit_index[::-1]
for i in range(0,pop_size):
new_pop[i,:] = pop[fit_index[i],:] #给最好的
new_fitvalue[i,:] = fitvalue[fit_index[i],:] #更新最好的 下标
return new_pop,new_fitvalue
8.获取最佳个体
def bestFit(pop,fitvalue):
fit_index = np.argsort(fitvalue,axis = 0)
fit_index = fit_index[::-1] #这里和selectParent的原因一样
best_one = pop[fit_index[0]]
best_fit = calAckLey(best_one)
return best_one,best_fit
9.我的测试函数
pop = create_pop(pop_size,dim,bounds)
pop_value = calAckLey(pop)
pop_fit = calFitvalue(pop_value)
print(pop_fit)
for i in range(0,100):
pop_pa = selectParent(pop,pop_fit) #选父代
pop_cross = crossover(pop_pa,cross_rate) #杂交
new_pop = np.concatenate((pop_cross,pop),axis = 0) #杂交后合并
pop_mut = mutation(new_pop,mutation_rate,bounds) #变异
new_pop = np.concatenate((pop_mut,new_pop),axis = 0)
new_pop_value = calAckLey(new_pop)
fitvalue = calFitvalue(new_pop_value)
pop,fitvalue = updatePop(new_pop,pop_size,fitvalue)
# 最低点到这,之后的没必要看
best_one,best_fit = bestFit(pop,fitvalue)
print(best_fit[0,0])
10.完整代码
PS:这个里面有些没用的注释懒得删了,凑合看吧。
```python
import numpy as np
import matplotlib.pyplot as plt
import math
from numpy.random.mtrand import f
dim = 10 # 个体取值维度,列
dna_size = 3 # DNA length
xbits = 64 # 与DNA长度相同
pop_size = 1 # 种群长度,行
cross_rate = 0.8 # 交叉(配)概率
mutation_rate = 0.003 # 编译(混合)概率
n_generations = 200 # 迭代次数
bounds = np.ones((dim,2),int) # 范围
bounds[:,0] = -15
bounds[:,1] = 30
def ackley(x,n): #这里的n就得用dim
a = 20
b = 0.2
# n = 2
c = 2*math.pi
sum1 = 0
sum2 = 0
for i in range(0,n):
sum1 = sum1 + x[i]**2
sum2 = sum2 + math.cos(c * x[i])
y = -a * math.exp(-b * np.sqrt(1/n * sum1)) - math.exp(1/n * sum2) + a + math.exp(1)
return y
def create_pop(pop_size,dim,bounds):
pop = np.random.rand(pop_size,dim) #生成一个随机的pop_size行dim列矩阵
for i in range(0,dim):
pop[:,i] = pop[:,i]*(bounds[i,1]-bounds[i,0])+bounds[i,0] #对每一行进行赋值
#可改,每行每列乘个数罢了
return pop
'''
def translateFitvalue(fitvalue,direction):
#direction = 1 translate
#direction = -1 n_translate
if direction == 1 :
fitvalue =
return
'''
def calAckLey(pop):
pop_x = pop.shape[0] #获取pop的行数,即pop_size
pop_value = np.zeros((pop_x,1),np.float64) #生成全为0的矩阵,之所以只有一列,因为是求y值,只需一个
for i in range(0,pop_x):
pop_value[i,0] = ackley(pop[i,:],dim) #对每一列求值然后给到,这里的pop[i,:]是一行单独作为一个数组传入ackley函数计算
return pop_value
def calFitvalue(pop_value):
value = 1/(pop_value + 0.0001) #防止分母为0
max_value = max(value)
min_value = min(value)
fitvalue = (value - min_value)/(max_value - min_value + 0.001) #防止最大值最小值相同导致为0,ps:这个fit数组里必有一个数是0
return fitvalue
def selectParent(pop,fitvalue):
fit_index = np.argsort(fitvalue, axis =0) #按行从小到大排列,返回对应元素排之前的下标
fit_index = fit_index[::-1] #此时是从大到小的坐标了
pop_x,pop_y = pop.shape #pop_x获取行数,pop_y获取列数,或许写成 pop_x = pop.shape[0] pop_y = pop.shape[1]更直观一点?
parent_num = int(pop_x//1.2) #//表示向下取整的最大数 ,不是所有人都可以交配的拜托
new_pop = np.zeros((parent_num,pop_y),np.float64) #列数不变,但新的pop行数小了,也就是个体少了
for i in range(0,parent_num):
new_pop[i,:] = pop[fit_index[i],:] #直接让子代成为父母中最优的那几个
return new_pop
def crossover(pop,cross_rate):
pop_x,pop_y = pop.shape
child_pop = np.zeros((2,pop_y),dtype = np.float64)
new_pop = np.zeros((1,pop_y),dtype = np.float64)
for i in range(0,pop_x):
if np.random.rand() < cross_rate : #落在cross_rate范围内就交配了
parent_pop1 = pop[i,:]
parent_pop2 = pop[np.random.randint(pop_x),:] # 有可能选到自己哦
n = np.random.rand() #随机截取父母长度
child_pop[0,:] = parent_pop1*n + parent_pop2*(1-n)
child_pop[1,:] = parent_pop1*(1-n) + parent_pop2*n
new_pop = np.concatenate((new_pop,child_pop),axis = 0) #其实直接输出就行,但返回的是new_pop所以得删掉首行的0
new_pop = np.delete(new_pop,0,axis = 0) #删掉第一行全0行
return new_pop
def mutation(pop,mutation_rate,bounds):#注意,pop或者mutation_rate要设好,自己测试的时候可以设大一点,不然样本少了就啥都没
pop_x,pop_y = pop.shape
new_pop = np.zeros((1,pop_y),dtype = np.float64)
for i in range(0,pop_x):
if np.random.rand() < mutation_rate:
new_pop_m = pop[i,:] #取出第i行
for j in range(0,pop_y):
if np.random.rand() < mutation_rate:
new_pop_m[j] = np.random.rand()*(bounds[j,1] - bounds[j,0]) + bounds[j,0] #第i行j个元素变异,方法和create_pop的方法一样
new_pop = np.append(new_pop,[new_pop_m],axis = 0)
new_pop = np.delete(new_pop,0,axis = 0)
return new_pop
def updatePop(pop,pop_size,fitvalue):
pop_x,pop_y = pop.shape
if pop_x < pop_size: #比之前的小,代表没把原来的pop和变异pop合起来
return pop
new_pop = np.zeros((pop_size,pop_y),dtype = np.float64)
new_fitvalue = np.zeros((pop_size,1),dtype = np.float64)
fit_index = np.argsort(fitvalue,axis = 0)
fit_index = fit_index[::-1]
for i in range(0,pop_size):
new_pop[i,:] = pop[fit_index[i],:] #给最好的
new_fitvalue[i,:] = fitvalue[fit_index[i],:] #更新最好的 下标
return new_pop,new_fitvalue
def bestFit(pop,fitvalue):
fit_index = np.argsort(fitvalue,axis = 0)
fit_index = fit_index[::-1]
best_one = pop[fit_index[0]]
best_fit = calAckLey(best_one)
return best_one,best_fit
pop = create_pop(pop_size,dim,bounds)
pop_value = calAckLey(pop)
pop_fit = calFitvalue(pop_value)
print(pop_fit)
for i in range(0,100):
pop_pa = selectParent(pop,pop_fit) #选父代
pop_cross = crossover(pop_pa,cross_rate) #杂交
new_pop = np.concatenate((pop_cross,pop),axis = 0) #杂交后合并
pop_mut = mutation(new_pop,mutation_rate,bounds) #变异
new_pop = np.concatenate((pop_mut,new_pop),axis = 0)
new_pop_value = calAckLey(new_pop)
fitvalue = calFitvalue(new_pop_value)
pop,fitvalue = updatePop(new_pop,pop_size,fitvalue)
# 最低点到这,之后的没必要看
best_one,best_fit = bestFit(pop,fitvalue)
print(best_fit[0,0])
'''
print("pop")
print(pop)
print("pop_value")
print(pop_value)
print("pop_fit")
print(pop_fit)
print("parent")
print(pop_pa)
print("crossover")
print(pop_cross)
print("mutation")
print(pop_mut)
'''
三、我的代码及解读
这里我们老师给的测试函数是:
"for GA"
def achley(X): #老师给的只要2种特征,选最大最小值在selecctParent(),fit_index = fit_index[::-1]不注释就是最小值,注释就找最大值
x = X[0]
y = X[1]
return -20.0 * np.exp(-0.2 * np.sqrt(0.5 * (x ** 2 + y ** 2))) \
- np.exp(0.5 * (np.cos(2 * np.pi * x) + np.cos(2 * np.pi * y))) \
+ np.exp(1) + 20
def rastrigin(x,y):
return 20+ x**2-10*np.cos(2*np.pi*x)+y**2-10*np.cos(2*np.pi*y)
同时要求:
算法函数的定义
def GA(func,xrange,xbits ,xdim):
其中: func 需要优化的函数
xrange 自变量的范围
xbits 二进制字符串数量
xdim 自变量的维度
1.创建种群
这里根据要求,需要生成特征值(基因型)为01字符串的种群。
思路是通过创建一个符合要求(行、列)的空的str类型的数组(这里需设置dtype = np.dtype(‘U2048’)才行,U后的数字是可以存储的最大长度,可以自设)
def createpop(size,dim,xbits,bounds):
#xbits 上限2048
pop = np.empty((size,dim),dtype = np.dtype('U2048')) #这里设置接收的字符长度
global dis
dis = (bounds[1] - bounds[0]) / (2**xbits)
global lowc
lowc = bounds[0]
global highc
highc = bounds[1]
#这里的dis,lowc,highc,是为了之后的二转十进制用的
for s in range(0,size):
for d in range(0,dim):
stre=""
for xb in range(0,xbits):
stre += str(random.randint(0,1))
pop[s][d]=stre
return pop
2.2转10进制
这一种转换方法非常巧妙,通过之前将特征值的上界和下界存储到highc和lowc里再用dis = (bounds[1] - bounds[0]) / (2**xbits)算出一个小区间的值,这样再通过yy = arr10 * dis + lowc进行转换,精度几乎不会丢失。
def DtoB(pop):#2转10
pop_x,pop_y = pop.shape
for x in range(0,pop_x):
for y in range(0,pop_y):
arr10 = int(pop[x][y],2)
yy = arr10 * dis + lowc
pop[x][y] = yy
return pop
硬转10进制
这样会导致精度丢失非常严重,大概当01字符串到64位以上就都丢了,10进制的话只能保证小数点后20位左右的精度
def DtoB(pop,xbits):
pop_x,pop_y = pop.shape
for x in range(0,pop_x):
for y in range(0,pop_y):
sum = 0
for b in range(0,xbits):
sum = sum + int(pop[x][y][b])*2**(xbits - b -1)
pop[x][y] = sum
return pop
3.10转2进制(非必要)
因为这里还是会丢精度,这倒不是算法的原因,这是因为计算机的浮点数精度的问题。所以为了解决这个问题,我后面通过深复制存了初始生成的种群,从而防止精度丢失。
def BtoD(pop): #10转2 , 这里没用上,最后为了保证精度,我用深复制存了及格数组
pop_x,pop_y = pop.shape
newpop = np.empty((pop_x,pop_y),dtype = np.dtype('U2048'))
for x in range(0,pop_x):
for y in range(0,pop_y):
yy = (float(pop[x][y])- lowc) / dis
yy = bin(int(yy))
yy = yy[2:]
need0 = xbits -len(yy)
zero = ""
if need0 > 0:
for i in range(0,need0):
zero += "0"
yy = zero + yy
newpop[x][y] = yy
return newpop
4.计算函数值
def calfunc(pop,func): #计算函数值
pop_x = pop.shape[0] #获取pop的行数,即pop_size
pop_value = np.zeros((pop_x,1),np.float64) #生成全为0的矩阵,之所以只有一列,因为是求y值,只需一个
if(func == "achley"):
for i in range(0,pop_x):
pop_value[i,0] = achley([float(pop[i,0]),float(pop[i,1])]) #对每一列求值然后给到,这里的pop[i,:]是一行单独作为一个数组传入ackley函数计算
elif(func == "rastrigin"):
for i in range(0,pop_x):
pop_value[i,0] = rastrigin(float(pop[i,0]),float(pop[i,1]))
return pop_value
5.计算适应值
def calFitvalue(pop_value):#计算适应值
value = 1/(pop_value + 0.0001) #防止分母为0
max_value = max(value)
min_value = min(value)
fitvalue = (value - min_value)/(max_value - min_value + 0.001) #防止最大值最小值相同导致为0,ps:这个fit数组里必有一个数是0
return fitvalue
6.自然选择
注意这里的fit_index的方向,这里的方向会决定你选的是最大值还是最小值
def selectParent(pop,fitvalue,func):#选父辈
fit_index = np.argsort(fitvalue, axis =0) #按行从小到大排列,返回对应元素排之前的下标
fit_index = fit_index[::-1] #此时是从大到小的坐标了
pop_x,pop_y = pop.shape #pop_x获取行数,pop_y获取列数,或许写成 pop_x = pop.shape[0] pop_y = pop.shape[1]更直观一点?
parent_num = int(pop_x * 0.8) #//表示向下取整的最大数 ,不是所有人都可以交配的拜托,选了80%
if parent_num == 0 : #不能为0
parent_num = 1
new_pop = np.zeros((parent_num,pop_y),dtype = np.dtype('U2048')) #列数不变,但新的pop行数小了,也就是个体少了
for i in range(0,parent_num):
new_pop[i,:] = pop[fit_index[i],:] #直接让选的父代成为父母中最优的那几个
new_need_born = pop_x - parent_num # 需要生的孩子数
return new_pop,new_need_born
7.杂交
def crossover(pop,cross_rate,need_born):
pop_x,pop_y = pop.shape
if pop_x % 2 == 0 : #如果种群数能被2整除
for x in range(0,int(pop_x / 2)): #配对父母
for y in range(0,pop_y):
if np.random.rand() < cross_rate: #如果在cross_rate的范围里则交配
pos = random.randrange(0,xbits) #选择杂交位置
pop_first = str(pop[x][y]) #这里及以下是杂交
pop_second = str(pop[x+1][y])
tmp = pop_first[pos:]
pop_first = pop_first[:pos] + pop_second[pos:]
pop_second = pop_second[:pos] + tmp
pop[x][y] = pop_first
pop[x+1][y] = pop_second
else :
for x in range(0,int((pop_x - 1)/ 2)): #如果多了一个单身狗,他就被踢了,因为最后一个即使备选也是这个里面适应值最低的
for y in range(0,pop_y):
if np.random.rand() < cross_rate:
pos = random.randrange(0,xbits)
pop_first = str(pop[x][y])
pop_second = str(pop[x+1][y])
tmp = pop_first[pos:]
pop_first = pop_first[:pos] + pop_second[pos:]
pop_second = pop_second[:pos] + tmp
pop[x][y] = pop_first
pop[x+1][y] = pop_second
return pop[:need_born,] #这里是保证返回需要补足的子代数个子代
8.变异
def mutation(pop,mutation_rate):
pop_x,pop_y = pop.shape
for x in range(0,pop_x):
for y in range(0,pop_y):
if np.random.rand() < mutation_rate: #如果在mutation_rate的范围内
pos = random.randrange(0,xbits) #选择变异位置
tmp = str(pop[x][y])
if tmp[pos] == "1" : #变异
tmp = tmp[:pos] + "0" + tmp[pos+1:]
else :
tmp = tmp[:pos] + "1" + tmp[pos+1:]
pop[x][y] = tmp
return pop
9.最优选择
def bestFit(pop,fitvalue,func):
fit_index = np.argsort(fitvalue, axis =0) #按行从小到大排列,返回对应元素排之前的下标
fit_index = fit_index[::-1] #此时是从大到小的坐标了
print(pop[fit_index[0],:])
print("其函数值为:")
y = calfunc(DtoB(pop[fit_index[0],:]),func)
y = float(y)
print(y)
return y
10.测试函数
def GA(func,xrange,xbits,xdim):
global bounds
bounds = xrange
global dim
dim = xdim
pop = createpop(pop_size,dim,int(xbits),bounds)
copypop = copy.deepcopy(pop) #记住要深复制!!!
pop_10 = DtoB(copypop)
pop_value = calfunc(pop_10,func)
pop_fit = calFitvalue(pop_value)
y = [] #用来存最佳y值
for g in range(0,n_generations):
pop_pa,need_born = selectParent(pop,pop_fit,func)
copypop_pa = copy.deepcopy(pop_pa)
cross_pop = crossover(copypop_pa,cross_rate,need_born)
new_pop = np.concatenate((pop_pa,cross_pop))
mutation_pop = mutation(new_pop,mutation_rate)
pop = copy.deepcopy(mutation_pop)
copypop = copy.deepcopy(pop) #记住要深复制!!!
pop_10 = DtoB(copypop)
pop_value = calfunc(pop_10,func)
pop_fit = calFitvalue(pop_value)
print('第{}代最优基因型是'.format(g+1))
y.append(bestFit(pop,pop_fit,func))
return y
if __name__=='__main__':
plt.rcParams['font.sans-serif']=['SimHei'] # 用来正常显示中文标签
plt.rcParams['axes.unicode_minus'] =False #负号显示
r_min, r_max = -32.768, 32.768
xaxis = np.arange(r_min, r_max, 2.0)
yaxis = np.arange(r_min, r_max, 2.0)
x, y = np.meshgrid(xaxis, yaxis)
results = achley([x, y])
figure = plt.figure()
axis = figure.gca( projection='3d')
axis.plot_surface(x, y, results, cmap='jet', shade= "false")
plt.title("achley函数的图像为:")
plt.show()
plt.contour(x, y, results)
plt.title("achley函数的等高线图为")
plt.show()
y = GA("achley",[-3,3],64,2)
plt.plot(y)
plt.title("最优解的的变化")
plt.show()
#==========================rastrigin==================
r_min, r_max = -5, 5
xaxis = np.arange(r_min, r_max, 0.1)
yaxis = np.arange(r_min, r_max, 0.1)
x, y = np.meshgrid(xaxis, yaxis)
results1 = rastrigin(x, y)
figure = plt.figure()
axis = figure.gca(projection='3d')
axis.plot_surface(x, y, results1, cmap='jet', shade="false")
plt.title("rastrigin函数的图像为:")
plt.show()
plt.contour(x, y, results1)
plt.title("rastrigin函数的等高线图为:")
plt.show()
y = GA("rastrigin",[-30,30],64,2)
plt.plot(y)
plt.title("最优解的的变化")
plt.show()
11.完整代码
import numpy as np
import random
import copy
from matplotlib import pyplot as plt
from scipy.misc import derivative
dim = 2 # 个体取值维度,列
xbits = 64 # 与DNA长度相同
pop_size = 1000 # 种群长度,行
cross_rate = 0.8 # 交叉(配)概率
mutation_rate = 0.003 # 编译(混合)概率
n_generations = 1000 # 迭代次数
bounds = [-3,3]
dis = 0
lowc = 0.
highc = 0.
def achley(X): #老师给的只要2种特征,选最大最小值在selecctParent(),fit_index = fit_index[::-1]不注释就是最小值,注释就找最大值
x = X[0]
y = X[1]
return -20.0 * np.exp(-0.2 * np.sqrt(0.5 * (x ** 2 + y ** 2))) \
- np.exp(0.5 * (np.cos(2 * np.pi * x) + np.cos(2 * np.pi * y))) \
+ np.exp(1) + 20
def rastrigin(x,y):
return 20+ x**2-10*np.cos(2*np.pi*x)+y**2-10*np.cos(2*np.pi*y)
def mhumps(x):
return abs(-1 / ((x - 0.3) ** 2 + 0.01)
+ 1 / ((x - 0.9) ** 2 + 0.04) - 6)
def createpop(size,dim,xbits,bounds):
#xbits 上限2048
pop = np.empty((size,dim),dtype = np.dtype('U2048')) #这里设置接收的字符长度
global dis
dis = (bounds[1] - bounds[0]) / (2**xbits)
global lowc
lowc = bounds[0]
global highc
highc = bounds[1]
#这里的dis,lowc,highc,是为了之后的二转十进制用的
for s in range(0,size):
for d in range(0,dim):
stre=""
for xb in range(0,xbits):
stre += str(random.randint(0,1))
pop[s][d]=stre
return pop
def DtoB(pop):#2转10
pop_x,pop_y = pop.shape
for x in range(0,pop_x):
for y in range(0,pop_y):
arr10 = int(pop[x][y],2)
yy = arr10 * dis + lowc
pop[x][y] = yy
return pop
def BtoD(pop): #10转2 , 这里没用上,最后为了保证精度,我用深复制存了及格数组
pop_x,pop_y = pop.shape
newpop = np.empty((pop_x,pop_y),dtype = np.dtype('U2048'))
for x in range(0,pop_x):
for y in range(0,pop_y):
yy = (float(pop[x][y])- lowc) / dis
yy = bin(int(yy))
yy = yy[2:]
need0 = xbits -len(yy)
zero = ""
if need0 > 0:
for i in range(0,need0):
zero += "0"
yy = zero + yy
newpop[x][y] = yy
return newpop
def calfunc(pop,func): #计算函数值
pop_x = pop.shape[0] #获取pop的行数,即pop_size
pop_value = np.zeros((pop_x,1),np.float64) #生成全为0的矩阵,之所以只有一列,因为是求y值,只需一个
if(func == "achley"):
for i in range(0,pop_x):
pop_value[i,0] = achley([float(pop[i,0]),float(pop[i,1])]) #对每一列求值然后给到,这里的pop[i,:]是一行单独作为一个数组传入ackley函数计算
elif(func == "rastrigin"):
for i in range(0,pop_x):
pop_value[i,0] = rastrigin(float(pop[i,0]),float(pop[i,1]))
return pop_value
def calFitvalue(pop_value):#计算适应值
value = 1/(pop_value + 0.0001) #防止分母为0
max_value = max(value)
min_value = min(value)
fitvalue = (value - min_value)/(max_value - min_value + 0.001) #防止最大值最小值相同导致为0,ps:这个fit数组里必有一个数是0
return fitvalue
def selectParent(pop,fitvalue,func):#选父辈
fit_index = np.argsort(fitvalue, axis =0) #按行从小到大排列,返回对应元素排之前的下标
fit_index = fit_index[::-1] #此时是从大到小的坐标了
pop_x,pop_y = pop.shape #pop_x获取行数,pop_y获取列数,或许写成 pop_x = pop.shape[0] pop_y = pop.shape[1]更直观一点?
parent_num = int(pop_x * 0.8) #//表示向下取整的最大数 ,不是所有人都可以交配的拜托,选了80%
if parent_num == 0 : #不能为0
parent_num = 1
new_pop = np.zeros((parent_num,pop_y),dtype = np.dtype('U2048')) #列数不变,但新的pop行数小了,也就是个体少了
for i in range(0,parent_num):
new_pop[i,:] = pop[fit_index[i],:] #直接让选的父代成为父母中最优的那几个
new_need_born = pop_x - parent_num # 需要生的孩子数
return new_pop,new_need_born
def crossover(pop,cross_rate,need_born):
pop_x,pop_y = pop.shape
if pop_x % 2 == 0 : #如果种群数能被2整除
for x in range(0,int(pop_x / 2)): #配对父母
for y in range(0,pop_y):
if np.random.rand() < cross_rate: #如果在cross_rate的范围里则交配
pos = random.randrange(0,xbits) #选择杂交位置
pop_first = str(pop[x][y]) #这里及以下是杂交
pop_second = str(pop[x+1][y])
tmp = pop_first[pos:]
pop_first = pop_first[:pos] + pop_second[pos:]
pop_second = pop_second[:pos] + tmp
pop[x][y] = pop_first
pop[x+1][y] = pop_second
else :
for x in range(0,int((pop_x - 1)/ 2)): #如果多了一个单身狗,他就被踢了,因为最后一个即使备选也是这个里面适应值最低的
for y in range(0,pop_y):
if np.random.rand() < cross_rate:
pos = random.randrange(0,xbits)
pop_first = str(pop[x][y])
pop_second = str(pop[x+1][y])
tmp = pop_first[pos:]
pop_first = pop_first[:pos] + pop_second[pos:]
pop_second = pop_second[:pos] + tmp
pop[x][y] = pop_first
pop[x+1][y] = pop_second
return pop[:need_born,] #这里是保证返回需要补足的子代数个子代
def mutation(pop,mutation_rate):
pop_x,pop_y = pop.shape
for x in range(0,pop_x):
for y in range(0,pop_y):
if np.random.rand() < mutation_rate: #如果在mutation_rate的范围内
pos = random.randrange(0,xbits) #选择变异位置
tmp = str(pop[x][y])
if tmp[pos] == "1" : #变异
tmp = tmp[:pos] + "0" + tmp[pos+1:]
else :
tmp = tmp[:pos] + "1" + tmp[pos+1:]
pop[x][y] = tmp
return pop
def bestFit(pop,fitvalue,func):
fit_index = np.argsort(fitvalue, axis =0) #按行从小到大排列,返回对应元素排之前的下标
fit_index = fit_index[::-1] #此时是从大到小的坐标了
print(pop[fit_index[0],:])
print("其函数值为:")
y = calfunc(DtoB(pop[fit_index[0],:]),func)
y = float(y)
print(y)
return y
def GA(func,xrange,xbits,xdim):
global bounds
bounds = xrange
global dim
dim = xdim
pop = createpop(pop_size,dim,int(xbits),bounds)
copypop = copy.deepcopy(pop) #记住要深复制!!!
pop_10 = DtoB(copypop)
pop_value = calfunc(pop_10,func)
pop_fit = calFitvalue(pop_value)
y = [] #用来存最佳y值
for g in range(0,n_generations):
pop_pa,need_born = selectParent(pop,pop_fit,func)
copypop_pa = copy.deepcopy(pop_pa)
cross_pop = crossover(copypop_pa,cross_rate,need_born)
new_pop = np.concatenate((pop_pa,cross_pop))
mutation_pop = mutation(new_pop,mutation_rate)
pop = copy.deepcopy(mutation_pop)
copypop = copy.deepcopy(pop) #记住要深复制!!!
pop_10 = DtoB(copypop)
pop_value = calfunc(pop_10,func)
pop_fit = calFitvalue(pop_value)
print('第{}代最优基因型是'.format(g+1))
y.append(bestFit(pop,pop_fit,func))
return y
if __name__=='__main__':
plt.rcParams['font.sans-serif']=['SimHei'] # 用来正常显示中文标签
plt.rcParams['axes.unicode_minus'] =False #负号显示
#=========GA===============
r_min, r_max = -32.768, 32.768
xaxis = np.arange(r_min, r_max, 2.0)
yaxis = np.arange(r_min, r_max, 2.0)
x, y = np.meshgrid(xaxis, yaxis)
results = achley([x, y])
figure = plt.figure()
axis = figure.gca( projection='3d')
axis.plot_surface(x, y, results, cmap='jet', shade= "false")
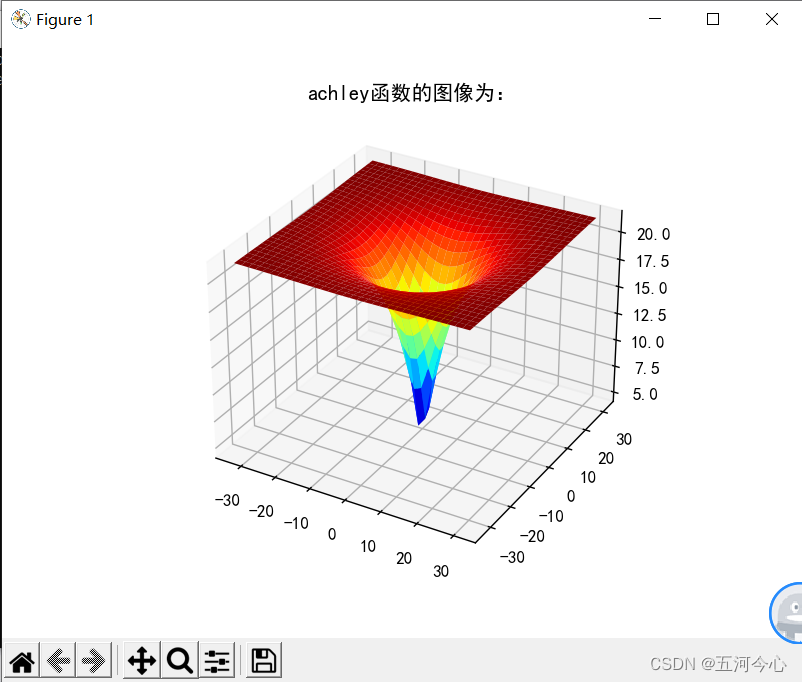
plt.title("achley函数的图像为:")
plt.show()
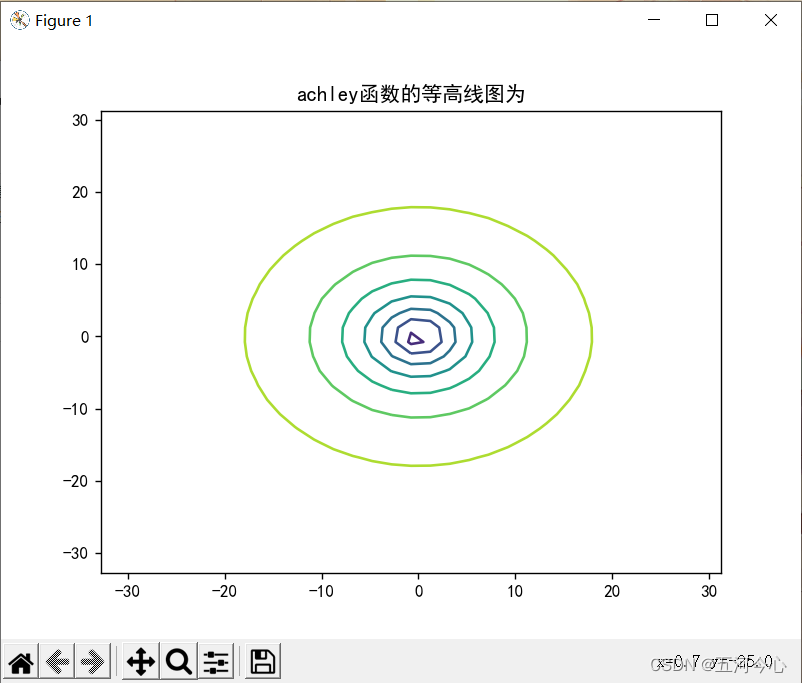
plt.contour(x, y, results)
plt.title("achley函数的等高线图为")
plt.show()
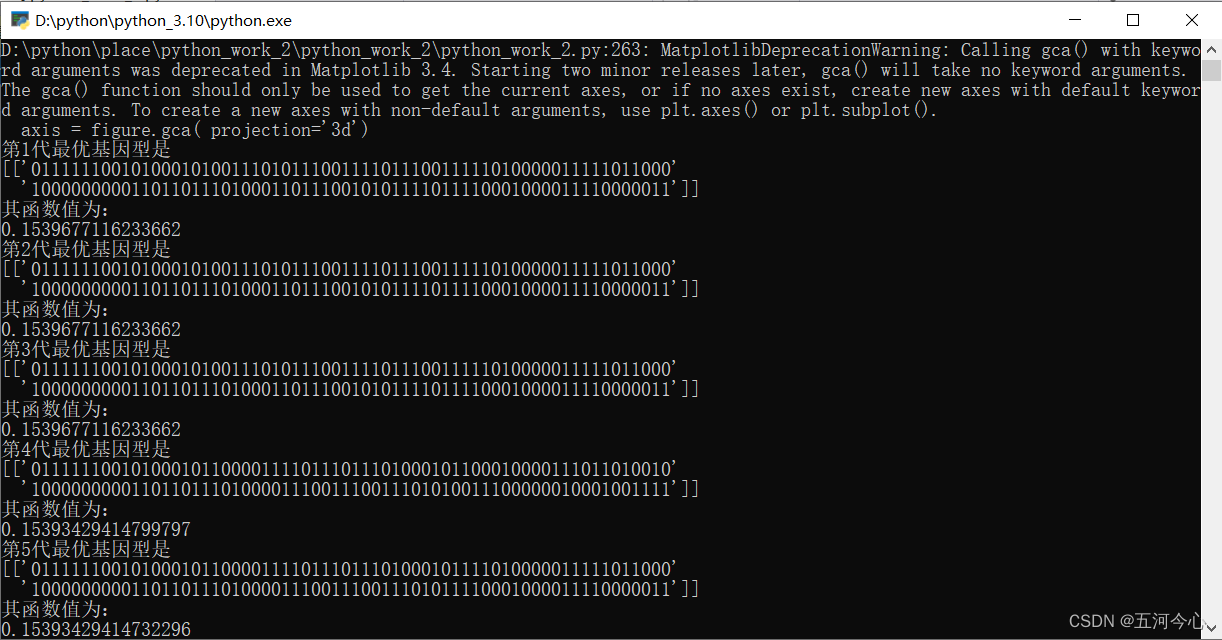
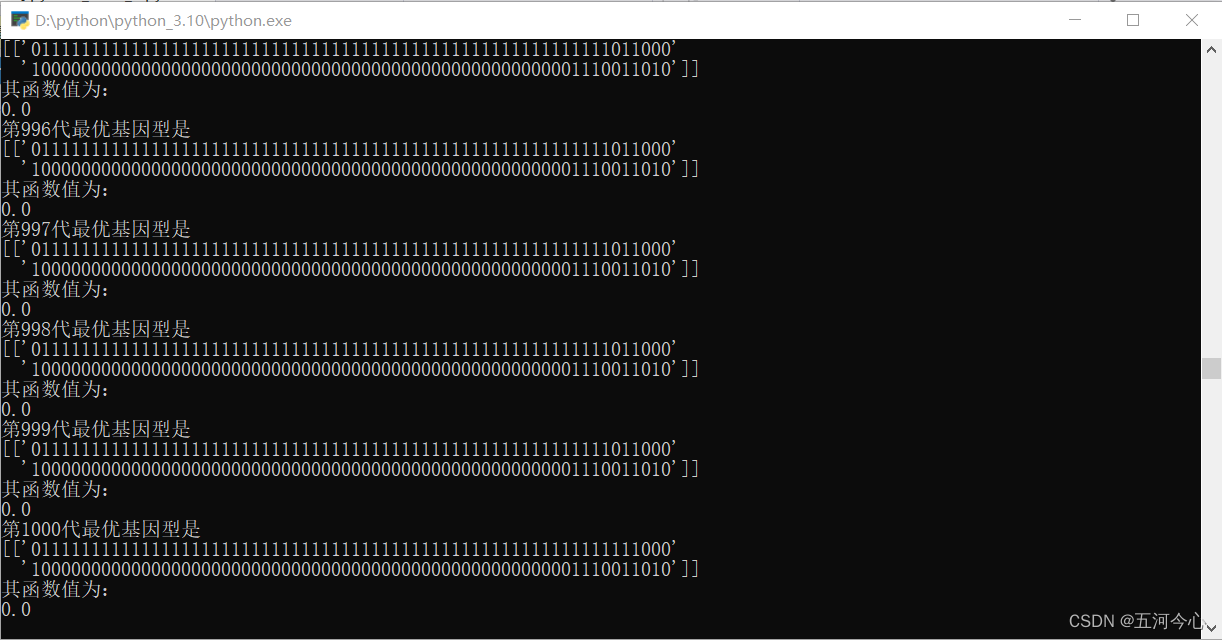
y = GA("achley",[-3,3],64,2)
plt.plot(y)
plt.title("最优解的的变化")
plt.show()
#==========================rastrigin==================
r_min, r_max = -5, 5
xaxis = np.arange(r_min, r_max, 0.1)
yaxis = np.arange(r_min, r_max, 0.1)
x, y = np.meshgrid(xaxis, yaxis)
results1 = rastrigin(x, y)
figure = plt.figure()
axis = figure.gca(projection='3d')
axis.plot_surface(x, y, results1, cmap='jet', shade="false")
plt.title("rastrigin函数的图像为:")
plt.show()
plt.contour(x, y, results1)
plt.title("rastrigin函数的等高线图为:")
plt.show()
y = GA("rastrigin",[-30,30],64,2)
plt.plot(y)
plt.title("最优解的的变化")
plt.show()
12.运行结果
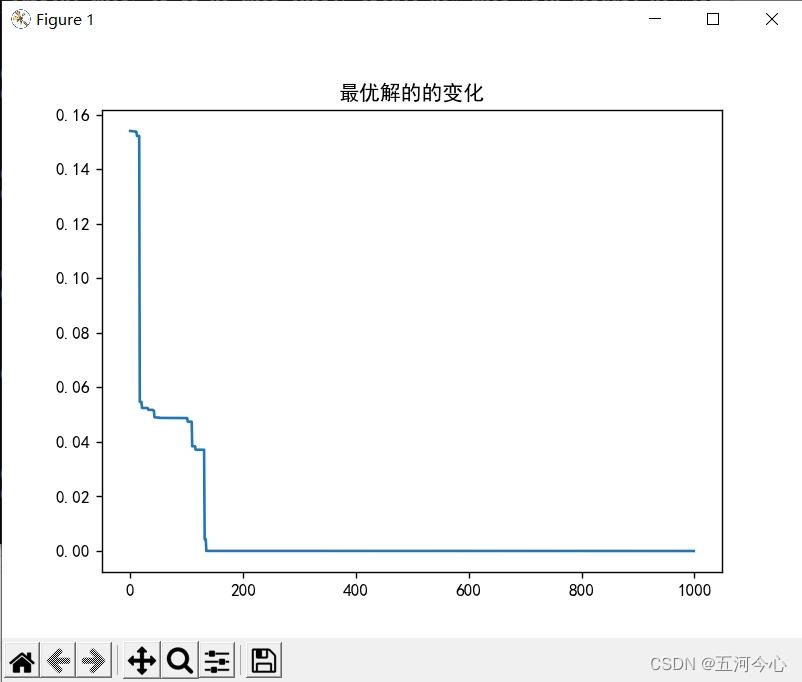
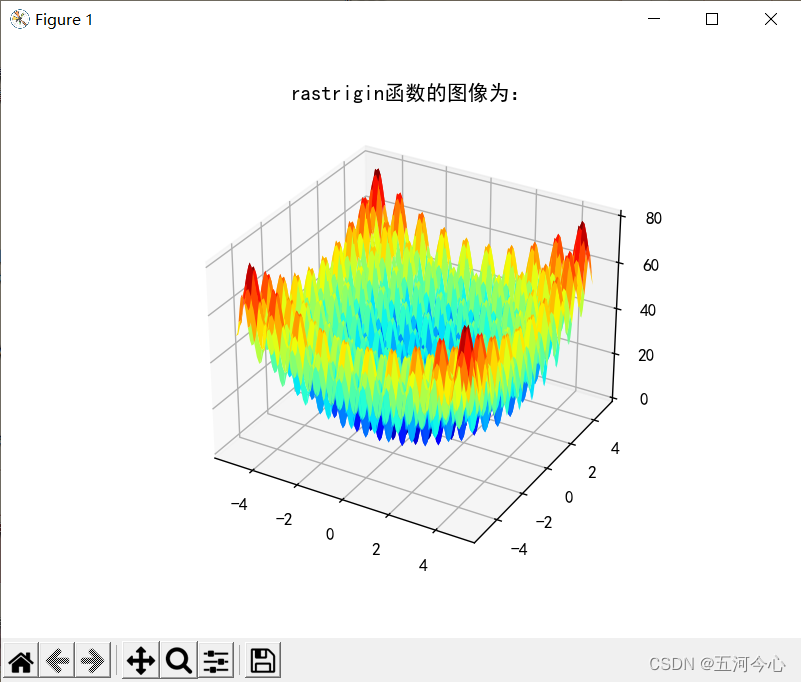
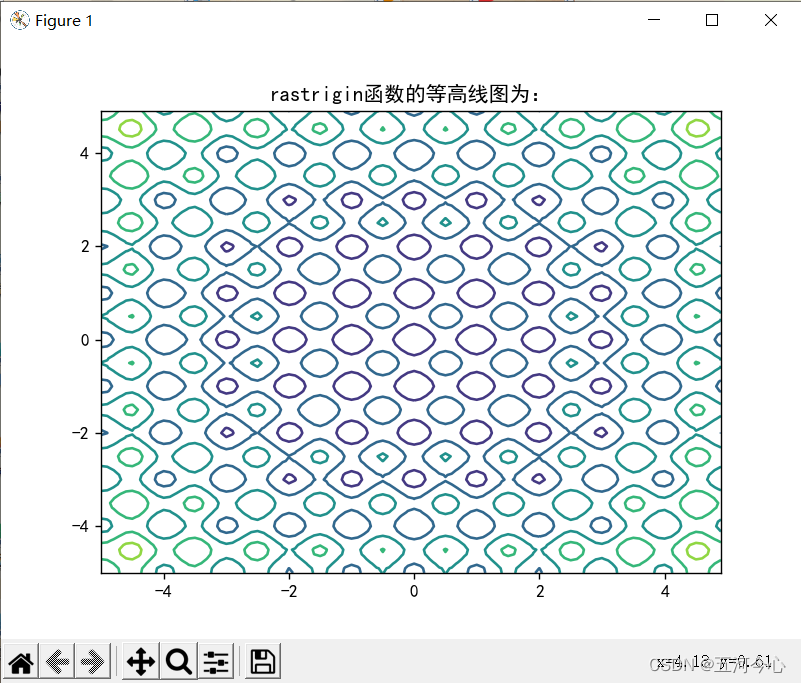
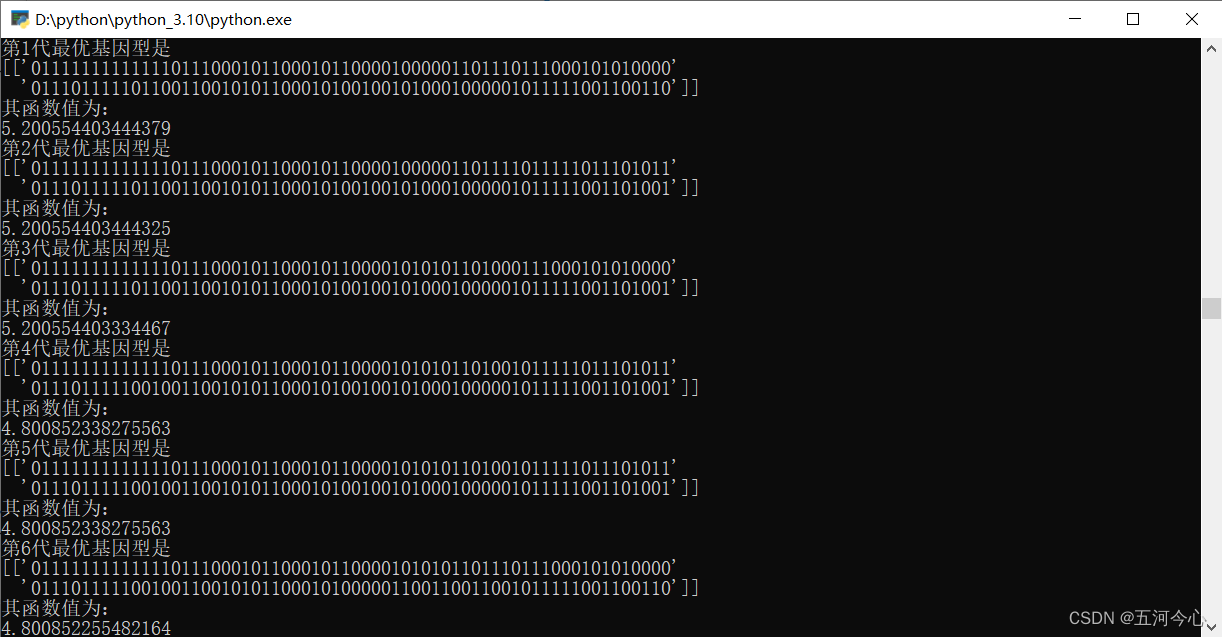
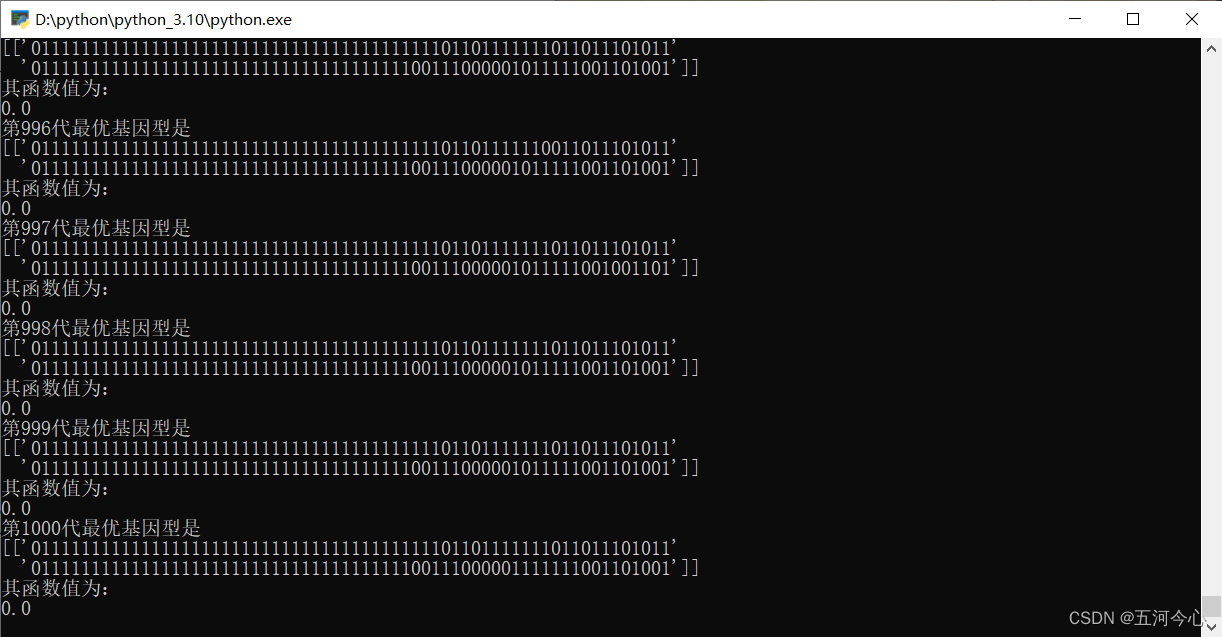
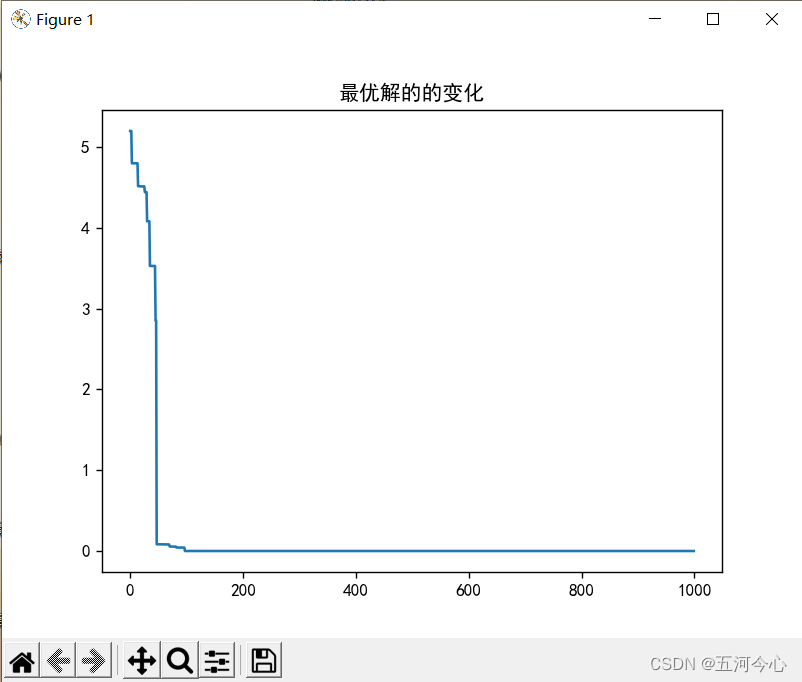
总结(非常重要,「请」务必先看)
在这一周多的时间里,我遇到了python的各种各样的奇怪bug(估计是因为我太菜了),其次通过这里关于GA算法的学习,我对它的每一个步骤都了如指掌。所以,看了这篇博客的你,请你在完成自己的作业时,千万不要直接抄了了事,因为这种东西,终归还是自己走一遍来的舒服。所谓“纸上得来终觉浅,绝知此事要躬行。”
诸君,努力吧。























 1万+
1万+











 被折叠的 条评论
为什么被折叠?
被折叠的 条评论
为什么被折叠?








