Assignment #4
Spectral Clustering
implement spectral.m
“opt”is used to accelerate the process of computing the eigenvalue and eigenvector, in this case the matrix is not symmetric.
function idx = spectral(W, k)
%SPECTRUAL spectral clustering
% Input:
% W: Adjacency matrix, N-by-N matrix
% k: number of clusters
% Output:
% idx: data point cluster labels, n-by-1 vector.
% YOUR CODE HERE
D = diag(sum(W));
L = D - W;
opt = struct('isreal', true);
[V,~] = eigs(L,D,k,'sm',opt);
idx = kmeans(V, k);
endimplement knn.graph
function W = knn_graph(X, k, threshold)
%KNN_GRAPH Construct W using KNN graph
% Input: X - data point features, n-by-p maxtirx.
% k - number of nn.
% threshold - distance threshold.
%
% Output:W - adjacency matrix, n-by-n matrix.
% YOUR CODE HERE
[n,~] = size(X);
D = EuDist2(X,X);
D(D == 0) = inf;
D(D > threshold) = inf;
[sD,idx] = sort(D,2);
sD(sD == inf) = 0;
sD = [sD(:,1:k), zeros(n,n-k)];
sD(sD ~= 0) = 1;
W = zeros(n,n);
for i = 1:size(idx,1)
W(i,idx(i,:)) = sD(i,:);
end
end
the spectral_exp1.m should be
load('cluster_data', 'X');
% Choose proper parameters
k_in_knn_graph = 500;
threshold = 0.3;
W = knn_graph(X, k_in_knn_graph, threshold);
idx = spectral(W, 2);
cluster_plot(X, idx);
figure;
idx = kmeans(X, 2);
cluster_plot(X, idx);
the result of spectral is:
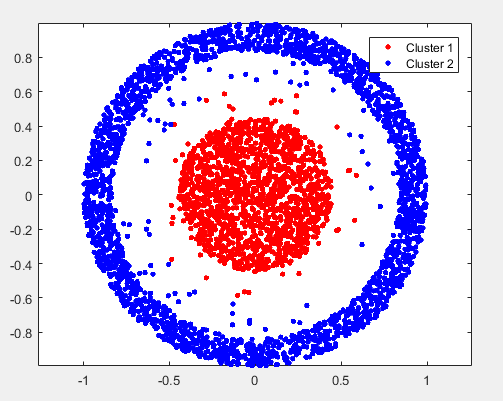
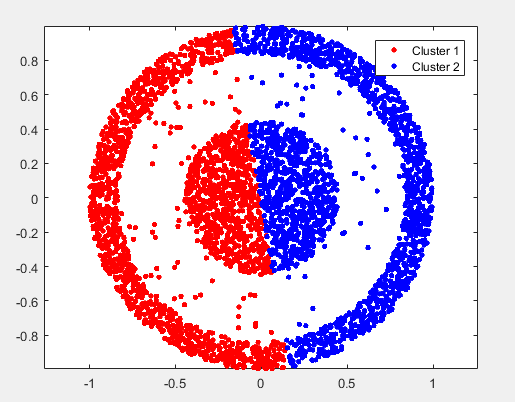
the result of kmeans is:
implement spectral_exp2.m:
load('TDT2_data', 'fea', 'gnd');
% YOUR CODE HERE
opt = struct('NeighborMode', 'KNN', 'k', 10, 'Weightmode', 'HeatKernel');
W = constructW(fea, opt);
accuracy_spectral = 0;
accuracy_kmeans = 0;
nmi_spectral = 0;
nmi_kmeans = 0;
nLoop = 500;
K = length(unique(gnd));
for i = 1 : nLoop
label = bestMap(gnd, spectral(W, K));
accuracy_spectral = accuracy_spectral + sum(gnd == label) / length(gnd) / nLoop;
nmi_spectral = nmi_spectral + MutualInfo(gnd, label) / nLoop;
label = bestMap(gnd, litekmeans(fea, K));
accuracy_kmeans = accuracy_kmeans + sum(gnd == label) / length(gnd) / nLoop;
nmi_kmeans = nmi_kmeans + MutualInfo(gnd, label) / nLoop;
end
fprintf('accuracy_spectral = %f, nmi_spectral = %f\n', accuracy_spectral, nmi_spectral);
fprintf('accuracy_kmeans = %f, nmi_kmeans = %f\n', accuracy_kmeans, nmi_kmeans);
| catagory | accuaracy | normalized mutual information |
|---|---|---|
| spectral | 0.800635 | 0.660020 |
| kmeans | 0.533962 | 0.317964 |
Principal Component Analysis
implement pca.m:
Though matlab provide the function, I suppose we should implement it all by ourself without call the pca.m or princomp.m given by matlab.
function [eigvector, eigvalue] = pca(data)
%PCA Principal Component Analysis
%
% Input:
% data - Data matrix. Each row vector of fea is a data point.
%
% Output:
% eigvector - Each column is an embedding function, for a new
% data point (row vector) x, y = x*eigvector
% will be the embedding result of x.
% eigvalue - The sorted eigvalue of PCA eigen-problem.
%
% YOUR CODE HERE
[N,~] = size(data);
X = data - repmat(mean(data),[N,1]);
sigma = 1/N * (X' * X);
[eigvector, eigvalue] = eigs(sigma, size(sigma,1) - 1);
endimplement hack_pca.m:
function img = hack_pca(filename)
% Input: filename -- input image file name/path
% Output: img -- image without rotation
img_r = double(imread(filename));
% YOUR CODE HERE
[r,c] = ind2sub(size(img_r),find(img_r ~= 255));
[W,~]=pca([r c]);
dir = W(:,1);
rot_angle = acos(dir(1)/norm(dir)) / pi * 180 - 90;
img = imrotate(img_r, rot_angle, 'bilinear', 'loose');
imshow(img,[]);
end| before | after |
|---|---|
 |  |
 |  |
 |  |
 |  |
 |  |
implement pca_exp1.m(in fact it is the code of (2)):
load('ORL_data', 'fea_Train', 'gnd_Train', 'fea_Test', 'gnd_Test');
% YOUR CODE HERE
% 1. Feature preprocessing
% 2. Run PCA
% 3. Visualize eigenface
% 4. Project data on to low dimensional space
% 5. Run KNN in low dimensional space
% 6. Recover face images form low dimensional space, visualize
%% (i)
[V,~] = pca(fea_Train);
show_face(V');
%% (ii)
[V,~] = pca(fea_Train);
reduced_dim = [8,16,32,64,128];
for i = 1:length(reduced_dim)
knn_model = fitcknn(fea_Train * V(:,1:reduced_dim(i)), gnd_Train);
label = predict(knn_model, fea_Test * V(:,1:reduced_dim(i)));
test_error = sum(label ~= gnd_Test) / length(gnd_Test);
fprintf('when reduced dimension is %d, test error = %f\n', reduced_dim(i), test_error);
end
%% (iii)
[V,~] = pca(fea_Train);
reduced_dim = [8,16,32,64,128];
figure;
show_face(fea_Train);
for i = 1:length(reduced_dim)
img = fea_Train * V(:,1:reduced_dim(i)) * pinv(V(:,1:reduced_dim(i)));
figure;
show_face(img);
endthe answer of (i) is :

the answer of (ii) is :
| reduced demension | test error |
|---|---|
| 8 | 0.245000 |
| 16 | 0.200000 |
| 32 | 0.180000 |
| 64 | 0.150000 |
| 128 | 0.150000 |
the answer of (iii) is :
origin

reduced dimensionality :8

reduced dimensionality :16

reduced dimensionality :32

reduced dimensionality :64

reduced dimensionality :128























 1万+
1万+

 被折叠的 条评论
为什么被折叠?
被折叠的 条评论
为什么被折叠?








