* 进行系统发育相关的分析:
phylotools_analysis(tree)
1. 数据可视化:
* 绘制系统发育树:
plot_tree(tree)
* 绘制节点度的分布图:
plot_degree(tree)
* 绘制节点高度的分布图:
plot_height(tree)
## github使用示例
library(“phytools”) # load the “phytools” package.
example<-read.csv(“example.splist.csv”,header=T) # read in the example species list.
phylo<-read.tree(“PhytoPhylo.tre”) # read in the megaphylogeny.
nodes<-read.csv(“nodes.csv”,header=T) # read in the nodes information of the megaphylogeny.
result<-S.PhyloMaker(spList=example, tree=phylo, nodes=nodes) # run the function S.PhyloMaker.
str(result) # the structure of the ouput of S.PhyloMaker.
par(mfrow=c(1,3),mar=c(0,0,1,0)) # show the phylogenies of the three scenarios.
plot(result
S
c
e
n
a
r
i
o
.
1
,
c
e
x
=
1.1
,
m
a
i
n
=
"
S
c
e
n
a
r
i
o
n
O
n
e
"
)
p
l
o
t
(
r
e
s
u
l
t
Scenario.1,cex=1.1,main="Scenarion One") plot(result
Scenario.1,cex=1.1,main="ScenarionOne")plot(resultScenario.2,cex=1.1,main=“Scenarion Two”)
plot(result$Scenario.3,cex=1.1,main=“Scenarion Three”)
## S.PhyloMaker完成分析脚本(有动手能力的可以改改):
S.PhyloMaker<-function (tree, spList, nodes, output.spList = T, scenarios = c(“S1”, “S2”, “S3”))
{
options(scipen=999)
tree0 <- tree
spList[sapply(spList, is.factor)] <- lapply(spList[sapply(spList, is.factor)], as.character)
if (any(duplicated(spListKaTeX parse error: Expected '}', got 'EOF' at end of input: … print(spListspecies[duplicated(spListKaTeX parse error: Expected 'EOF', got '}' at position 18: …ecies)]) }̲ spList <- …species), ]
spList.original <- spList
spList
s
p
e
c
i
e
s
<
−
g
s
u
b
(
"
"
,
"
"
,
s
p
L
i
s
t
species <- gsub(" ", "_", spList
species<−gsub("","",spListspecies)
spList
s
p
e
c
i
e
s
<
−
g
s
u
b
(
"
(
[
[
:
a
l
p
h
a
:
]
]
)
"
,
"
U
1
"
,
s
p
L
i
s
t
species <- gsub("(^[[:alpha:]])", "\\U\\1", spList
species<−gsub("([[:alpha:]])","U1",spListspecies, perl = TRUE)
spList
g
e
n
u
s
<
−
g
s
u
b
(
"
(
[
[
:
a
l
p
h
a
:
]
]
)
"
,
"
U
1
"
,
s
p
L
i
s
t
genus <- gsub("(^[[:alpha:]])", "\\U\\1", spList
genus<−gsub("([[:alpha:]])","U1",spListgenus, perl = TRUE)
spList
f
a
m
i
l
y
<
−
g
s
u
b
(
"
(
[
[
:
a
l
p
h
a
:
]
]
)
"
,
"
U
1
"
,
s
p
L
i
s
t
family <- gsub("(^[[:alpha:]])", "\\U\\1", spList
family<−gsub("([[:alpha:]])","U1",spListfamily, perl = TRUE)
rnN <- data.frame(node.label = paste(“N”, 1:length(tree
n
o
d
e
.
l
a
b
e
l
)
,
s
e
p
=
"
"
)
,
o
r
i
N
=
t
r
e
e
node.label), sep = ""), oriN = tree
node.label),sep=""),oriN=treenode.label, stringsAsFactors = FALSE)
nodes[,c(“level”,“family”,“genus”,“rn”,“bn”,“taxa”)]<-lapply(nodes[,c(“level”,“family”,“genus”,“rn”,“bn”,“taxa”)], as.character)
tree
n
o
d
e
.
l
a
b
e
l
<
−
p
a
s
t
e
(
"
N
"
,
1
:
l
e
n
g
t
h
(
t
r
e
e
node.label <- paste("N", 1:length(tree
node.label<−paste("N",1:length(treenode.label), sep = “”)
kk<-c()
for (i in 1:length(treeKaTeX parse error: Expected '}', got 'EOF' at end of input: …,substring(treetip.label[i],1,gregexpr(““,treeKaTeX parse error: Expected 'EOF', got '}' at position 31: …1]][1]-1)) }̲ m<-data.fr…tip.label)
m<-merge(m,nodes[,c(“genus”,“family”)])
mX <- m
m <- m[,c(“genus”,“family”)]
m <- m[!duplicated(m
g
e
n
u
s
)
,
]
d
i
m
n
a
m
e
s
(
m
)
[
[
2
]
]
[
2
]
<
−
"
f
a
m
i
l
y
i
n
P
h
y
t
o
P
h
y
l
o
"
m
<
−
m
[
,
c
(
"
g
e
n
u
s
"
,
"
f
a
m
i
l
y
i
n
P
h
y
t
o
P
h
y
l
o
"
)
]
m
0
<
−
s
p
L
i
s
t
[
!
d
u
p
l
i
c
a
t
e
d
(
s
p
L
i
s
t
genus),] dimnames(m)[[2]][2] <- "family_in_PhytoPhylo" m<-m[,c("genus","family_in_PhytoPhylo")] m0 <- spList[!duplicated(spList
genus),]dimnames(m)[[2]][2]<−"familyinPhytoPhylo"m<−m[,c("genus","familyinPhytoPhylo")]m0<−spList[!duplicated(spListgenus), c(“genus”,“family”)]
dimnames(m0)[[2]][2] <- “family_in_spList”
mm<-merge(m0, m)
g<-mm[which(is.na(match(paste(mm
g
e
n
u
s
,
m
m
genus,mm
genus,mmfamily_in_spList,sep=””),paste(mm
g
e
n
u
s
,
m
m
genus,mm
genus,mmfamily_in_PhytoPhylo,sep=“")))),]
if (dim(g)[1]>0)
{
print(“Taxonomic classification not consistent between spList and PhytoPhylo.”)
print(g)
}
add.tip <- spList[which(is.na(match(spList
s
p
e
c
i
e
s
,
t
r
e
e
species, tree
species,treetip.label))), ]
status <- rep(“match(prune)”, dim(spList)[1])
status[which(is.na(match(spList
s
p
e
c
i
e
s
,
t
r
e
e
species, tree
species,treetip.label)))] <- “match(add)”
if (dim(add.tip)[1] == 0 & length(na.omit(match(spList
s
p
e
c
i
e
s
,
t
r
e
e
species, tree
species,treetip.label))) == 0)
stop(“Incorrect format of species list.”)
if (length(setdiff(spList
s
p
e
c
i
e
s
,
t
r
e
e
0
species, tree0
species,tree0tip.label)) == 0 & length(na.omit(match(spList
s
p
e
c
i
e
s
,
t
r
e
e
species, tree
species,treetip.label))) > 0)
{
print(“There is no species needs to be added, all the species are pruned from PhytoPhylo.”)
splis <- spList.original
treeX <- drop.tip(tree0, setdiff(tree0
t
i
p
.
l
a
b
e
l
,
s
p
l
i
s
tip.label, splis
tip.label,splisspecies))
splis
s
t
a
t
u
s
<
−
"
m
a
t
c
h
(
p
r
u
n
e
)
"
p
h
y
l
o
0
<
−
l
i
s
t
(
S
c
e
n
a
r
i
o
.
1
=
N
U
L
L
,
S
c
e
n
a
r
i
o
.
2
=
N
U
L
L
,
S
c
e
n
a
r
i
o
.
3
=
N
U
L
L
,
S
p
e
c
i
e
s
.
l
i
s
t
=
s
p
l
i
s
)
i
f
(
"
S
1
"
status <- "match(prune)" phylo0 <- list(Scenario.1 = NULL, Scenario.2 = NULL, Scenario.3 = NULL, Species.list = splis) if ("S1" %in% scenarios) {phylo0
status<−"match(prune)"phylo0<−list(Scenario.1=NULL,Scenario.2=NULL,Scenario.3=NULL,Species.list=splis)if("S1"Scenario.1 <- treeX}
if (“S2” %in% scenarios) {phylo0KaTeX parse error: Expected 'EOF', got '}' at position 20: …ario.2 <- treeX}̲ if ("S…Scenario.3 <- treeX}
phylo0[sapply(phylo0, is.null)] <- NULL
return(phylo0)
stop()
}
add.tip
s
o
r
t
<
−
"
"
a
d
d
.
t
i
p
sort <- "" add.tip
sort<−""add.tipsort[which(!is.na(match(add.tip
g
e
n
u
s
,
n
o
d
e
s
[
n
o
d
e
s
genus, nodes[nodes
genus,nodes[nodeslevel == “G”, ]
g
e
n
u
s
)
)
)
]
<
−
"
G
1
"
a
d
d
.
t
i
p
genus)))] <- "G1" add.tip
genus)))]<−"G1"add.tipsort[which(is.na(match(add.tip
g
e
n
u
s
,
n
o
d
e
s
[
n
o
d
e
s
genus, nodes[nodes
genus,nodes[nodeslevel == “G”, ]KaTeX parse error: Expected 'EOF', got '&' at position 9: genus)) &̲ !is.na(match(a…family, nodes[nodes
l
e
v
e
l
=
=
"
F
"
,
]
level == "F", ]
level=="F",]family)))] <- “F1”
add.tip
s
o
r
t
[
a
d
d
.
t
i
p
sort[add.tip
sort[add.tipsort == “F1”][duplicated(add.tip[add.tip
s
o
r
t
=
=
"
F
1
"
,
]
sort == "F1", ]
sort=="F1",]genus)] <- “F2”
a <- which(add.tipKaTeX parse error: Expected '}', got 'EOF' at end of input: … print(add.tipspecies[a])
status[match(add.tip
s
p
e
c
i
e
s
[
a
]
,
s
p
L
i
s
t
species[a], spList
species[a],spListspecies)] <- “unmatch”
}
spList.original
s
t
a
t
u
s
<
−
s
t
a
t
u
s
i
f
(
"
S
1
"
t
1
<
−
t
r
e
e
r
n
N
1
<
−
r
n
N
n
G
<
−
n
o
d
e
s
[
n
o
d
e
s
status <- status if ("S1" %in% scenarios) { t1 <- tree rnN1<-rnN nG <- nodes[nodes
status<−statusif("S1"t1<−treernN1<−rnNnG<−nodes[nodeslevel == “G”, ]
nF <- nodes[nodes
l
e
v
e
l
=
=
"
F
"
,
]
d
a
t
a
<
−
a
d
d
.
t
i
p
[
a
d
d
.
t
i
p
level == "F", ] data <- add.tip[add.tip
level=="F",]data<−add.tip[add.tipsort == “F1”, ]
if (dim(data)[1] > 0) {
for (i in 1:dim(data)[1]) {
n <- match(data
f
a
m
i
l
y
[
i
]
,
n
F
family[i], nF
family[i],nFfamily)
g <- nF
g
e
n
.
n
[
n
]
s
<
−
n
F
gen.n[n] s <- nF
gen.n[n]s<−nFsp.n[n]
if (g == 1 & s == 1) {
num <- grep(nF
t
a
x
a
[
n
]
,
t
1
taxa[n], t1
taxa[n],t1tip.label)
len <- t1
e
d
g
e
.
l
e
n
g
t
h
[
m
a
t
c
h
(
n
u
m
,
t
1
edge.length[match(num, t1
edge.length[match(num,t1edge[, 2])]
t1 <- bind.tip(t1, tip.label = data
s
p
e
c
i
e
s
[
i
]
,
e
d
g
e
.
l
e
n
g
t
h
=
l
e
n
,
w
h
e
r
e
=
n
u
m
,
p
o
s
i
t
i
o
n
=
l
e
n
)
n
F
species[i], edge.length = len, where = num, position = len) nF
species[i],edge.length=len,where=num,position=len)nFgen.n[n] <- g + 1
nF
s
p
.
n
[
n
]
<
−
s
+
1
n
u
m
<
−
g
r
e
p
(
n
F
sp.n[n] <- s + 1 num <- grep(nF
sp.n[n]<−s+1num<−grep(nFtaxa[n], t1
t
i
p
.
l
a
b
e
l
)
t
1
tip.label) t1
tip.label)t1node.label[match(t1
e
d
g
e
[
m
a
t
c
h
(
n
u
m
,
t
1
edge[match(num, t1
edge[match(num,t1edge[, 2]), 1], unique(t1
e
d
g
e
[
,
1
]
)
)
]
<
−
p
a
s
t
e
(
"
N
N
"
,
t
1
edge[, 1]))] <- paste("NN", t1
edge[,1]))]<−paste("NN",t1Nnode + 1, sep = “”)
rnN1
n
o
d
e
.
l
a
b
e
l
[
m
a
t
c
h
(
n
F
node.label[match(nF
node.label[match(nFbn[n], rnN1
n
o
d
e
.
l
a
b
e
l
)
]
<
−
p
a
s
t
e
(
"
N
N
"
,
t
1
node.label)]<- paste("NN", t1
node.label)]<−paste("NN",t1Nnode + 1, sep = “”)
nF
b
n
[
n
]
<
−
p
a
s
t
e
(
"
N
N
"
,
t
1
bn[n] <- paste("NN", t1
bn[n]<−paste("NN",t1Nnode + 1, sep = “”)
nFKaTeX parse error: Expected 'EOF', got '}' at position 33: … }̲ …edge[, 1])[match(nF
b
n
[
n
]
,
t
1
bn[n], t1
bn[n],t1node.label)]
len <- nF
b
n
.
b
l
[
n
]
t
1
<
−
b
i
n
d
.
t
i
p
(
t
1
,
t
i
p
.
l
a
b
e
l
=
d
a
t
a
bn.bl[n] t1 <- bind.tip(t1, tip.label = data
bn.bl[n]t1<−bind.tip(t1,tip.label=dataspecies[i], edge.length = len, where = num)
}
}
}
data <- add.tip[add.tipKaTeX parse error: Expected '}', got 'EOF' at end of input: …grep(paste(datagenus[i], "”, sep = “”), t1KaTeX parse error: Expected '}', got 'EOF' at end of input: … len <- t1edge.length[match(num, t1
e
d
g
e
[
,
2
]
)
]
t
1
<
−
b
i
n
d
.
t
i
p
(
t
1
,
t
i
p
.
l
a
b
e
l
=
d
a
t
a
edge[, 2])] t1 <- bind.tip(t1, tip.label = data
edge[,2])]t1<−bind.tip(t1,tip.label=dataspecies[i], edge.length = len, where = num, position = len)
}
if (length(n) > 1) {
num <- fastMRCA(t1, t1
t
i
p
.
l
a
b
e
l
[
m
i
n
(
n
)
]
,
t
1
tip.label[min(n)], t1
tip.label[min(n)],t1tip.label[max(n)])
len <- fastDist(t1, t1
t
i
p
.
l
a
b
e
l
[
m
i
n
(
n
)
]
,
t
1
tip.label[min(n)], t1
tip.label[min(n)],t1tip.label[max(n)])/2
t1 <- bind.tip(t1, tip.label = dataKaTeX parse error: Expected 'EOF', got '}' at position 63: … }̲ …tip.label), which(!is.na(match(t1
t
i
p
.
l
a
b
e
l
,
s
p
L
i
s
t
tip.label, spList
tip.label,spListspecies))))
t1 <- drop.tip(t1, tip = toDrop)
Re <- which(!is.na(match(t1
n
o
d
e
.
l
a
b
e
l
,
r
n
N
1
node.label, rnN1
node.label,rnN1node.label)))
noRe <- which(is.na(match(t1
n
o
d
e
.
l
a
b
e
l
,
r
n
N
1
node.label, rnN1
node.label,rnN1node.label)))
t1
n
o
d
e
.
l
a
b
e
l
[
R
e
]
<
−
r
n
N
1
node.label[Re] <- rnN1
node.label[Re]<−rnN1oriN[match(t1
n
o
d
e
.
l
a
b
e
l
,
r
n
N
1
node.label, rnN1
node.label,rnN1node.label)[Re]]
t1KaTeX parse error: Expected 'EOF', got '}' at position 28: …oRe] <- "" }̲ else { …level == “G”, ]
nF <- nodes[nodes
l
e
v
e
l
=
=
"
F
"
,
]
d
a
t
a
<
−
a
d
d
.
t
i
p
[
a
d
d
.
t
i
p
level == "F", ] data <- add.tip[add.tip
level=="F",]data<−add.tip[add.tipsort == “F1”, ]
if (dim(data)[1] > 0) {
for (i in 1:dim(data)[1]) {
n <- match(data
f
a
m
i
l
y
[
i
]
,
n
F
family[i], nF
family[i],nFfamily)
g <- nF
g
e
n
.
n
[
n
]
s
<
−
n
F
gen.n[n] s <- nF
gen.n[n]s<−nFsp.n[n]
if (g == 1 & s == 1) {
num <- grep(nF
t
a
x
a
[
n
]
,
t
2
taxa[n], t2
taxa[n],t2tip.label)
len <- t2
e
d
g
e
.
l
e
n
g
t
h
[
m
a
t
c
h
(
n
u
m
,
t
2
edge.length[match(num, t2
edge.length[match(num,t2edge[,2])] * sample((1:99)/100,1)
t2 <- bind.tip(t2, tip.label = data
s
p
e
c
i
e
s
[
i
]
,
e
d
g
e
.
l
e
n
g
t
h
=
l
e
n
,
w
h
e
r
e
=
n
u
m
,
p
o
s
i
t
i
o
n
=
l
e
n
)
n
F
species[i], edge.length = len, where = num, position = len) nF
species[i],edge.length=len,where=num,position=len)nFgen.n[n] <- g + 1
nF
s
p
.
n
[
n
]
<
−
s
+
1
n
u
m
<
−
g
r
e
p
(
d
a
t
a
sp.n[n] <- s + 1 num <- grep(data
sp.n[n]<−s+1num<−grep(dataspecies[i], t2
t
i
p
.
l
a
b
e
l
)
t
2
tip.label) t2
tip.label)t2node.label[match(t2
e
d
g
e
[
m
a
t
c
h
(
n
u
m
,
t
2
edge[match(num, t2
edge[match(num,t2edge[, 2]), 1], unique(t2
e
d
g
e
[
,
1
]
)
)
]
<
−
p
a
s
t
e
(
"
N
N
"
,
t
2
edge[,1]))] <- paste("NN", t2
edge[,1]))]<−paste("NN",t2Nnode + 1, sep = “”)
rnN2
n
o
d
e
.
l
a
b
e
l
[
m
a
t
c
h
(
n
F
node.label[match(nF
node.label[match(nFbn[n], rnN2
n
o
d
e
.
l
a
b
e
l
)
]
<
−
p
a
s
t
e
(
"
N
N
"
,
t
2
node.label)]<- paste("NN", t2
node.label)]<−paste("NN",t2Nnode + 1, sep = “”)
nF
b
n
[
n
]
<
−
p
a
s
t
e
(
"
N
N
"
,
t
2
bn[n] <- paste("NN", t2
bn[n]<−paste("NN",t2Nnode + 1, sep = “”)
nFKaTeX parse error: Expected 'EOF', got '}' at position 33: … }̲ …edge[, 1])[match(nF
b
n
[
n
]
,
t
2
bn[n], t2
bn[n],t2node.label)]
len <- t2
e
d
g
e
.
l
e
n
g
t
h
[
m
a
t
c
h
(
n
u
m
,
t
2
edge.length[match(num,t2
edge.length[match(num,t2edge[,2])] * sample((1:99)/100,1)
t2 <- bind.tip(t2, tip.label = data
s
p
e
c
i
e
s
[
i
]
,
e
d
g
e
.
l
e
n
g
t
h
=
l
e
n
,
w
h
e
r
e
=
n
u
m
,
p
o
s
i
t
i
o
n
=
l
e
n
)
n
F
species[i], edge.length = len, where = num, position = len) nF
species[i],edge.length=len,where=num,position=len)nFgen.n[n] <- g + 1
nF
s
p
.
n
[
n
]
<
−
s
+
1
n
u
m
<
−
g
r
e
p
(
d
a
t
a
sp.n[n] <- s + 1 num <- grep(data
sp.n[n]<−s+1num<−grep(dataspecies[i], t2
t
i
p
.
l
a
b
e
l
)
t
2
tip.label) t2
tip.label)t2node.label[match(t2
e
d
g
e
[
m
a
t
c
h
(
n
u
m
,
t
2
edge[match(num, t2
edge[match(num,t2edge[, 2]), 1], unique(t2
e
d
g
e
[
,
1
]
)
)
]
<
−
p
a
s
t
e
(
"
N
N
"
,
t
2
edge[, 1]))] <- paste("NN", t2
edge[,1]))]<−paste("NN",t2Nnode + 1, sep = “”)
rnN2
n
o
d
e
.
l
a
b
e
l
[
m
a
t
c
h
(
n
F
node.label[match(nF
node.label[match(nFbn[n], rnN2
n
o
d
e
.
l
a
b
e
l
)
]
<
−
p
a
s
t
e
(
"
N
N
"
,
t
2
node.label)]<- paste("NN", t2
node.label)]<−paste("NN",t2Nnode + 1, sep = “”)
nF
b
n
[
n
]
<
−
p
a
s
t
e
(
"
N
N
"
,
t
2
bn[n] <- paste("NN", t2
bn[n]<−paste("NN",t2Nnode + 1, sep = “”)
nF
b
n
.
b
l
[
n
]
<
−
n
F
bn.bl[n] <- nF
bn.bl[n]<−nFbn.bl[n]+len
}
}
}
data <- add.tip[add.tipKaTeX parse error: Expected '}', got 'EOF' at end of input: …grep(paste(datagenus[i], “", sep = “”), t2KaTeX parse error: Expected '}', got 'EOF' at end of input: … len <- t2edge.length[match(num, t2
e
d
g
e
[
,
2
]
)
]
∗
s
a
m
p
l
e
(
(
1
:
99
)
/
100
,
1
)
t
2
<
−
b
i
n
d
.
t
i
p
(
t
2
,
t
i
p
.
l
a
b
e
l
=
d
a
t
a
edge[, 2])] * sample((1:99)/100,1) t2 <- bind.tip(t2, tip.label = data
edge[,2])]∗sample((1:99)/100,1)t2<−bind.tip(t2,tip.label=dataspecies[i], edge.length = len, where = num, position = len)
}
if (length(n) > 1) {
num <- sample(n,1)
len <- t2
e
d
g
e
.
l
e
n
g
t
h
[
m
a
t
c
h
(
n
u
m
,
t
2
edge.length[match(num, t2
edge.length[match(num,t2edge[, 2])] * sample((1:99)/100,1)
t2 <- bind.tip(t2, tip.label = dataKaTeX parse error: Expected 'EOF', got '}' at position 79: … }̲ …tip.label), which(!is.na(match(t2
t
i
p
.
l
a
b
e
l
,
s
p
L
i
s
t
tip.label, spList
tip.label,spListspecies))))
t2 <- drop.tip(t2, tip = toDrop)
Re <- which(!is.na(match(t2
n
o
d
e
.
l
a
b
e
l
,
r
n
N
2
node.label, rnN2
node.label,rnN2node.label)))
noRe <- which(is.na(match(t2
n
o
d
e
.
l
a
b
e
l
,
r
n
N
2
node.label, rnN2
node.label,rnN2node.label)))
t2
n
o
d
e
.
l
a
b
e
l
[
R
e
]
<
−
r
n
N
2
node.label[Re] <- rnN2
node.label[Re]<−rnN2oriN[match(t2
n
o
d
e
.
l
a
b
e
l
,
r
n
N
2
node.label, rnN2
node.label,rnN2node.label)[Re]]
t2KaTeX parse error: Expected 'EOF', got '}' at position 28: …oRe] <- "" }̲ else { …level == “G”, ]
nF <- nodes[nodes
l
e
v
e
l
=
=
"
F
"
,
]
d
a
t
a
<
−
a
d
d
.
t
i
p
[
a
d
d
.
t
i
p
level == "F", ] data <- add.tip[add.tip
level=="F",]data<−add.tip[add.tipsort == “F1”, ]
if (dim(data)[1] > 0) {
for (i in 1:dim(data)[1]) {
n <- match(data
f
a
m
i
l
y
[
i
]
,
n
F
family[i], nF
family[i],nFfamily)
g <- nF
g
e
n
.
n
[
n
]
s
<
−
n
F
gen.n[n] s <- nF
gen.n[n]s<−nFsp.n[n]
if (g == 1 & s == 1) {
num <- grep(nF
t
a
x
a
[
n
]
,
t
3
taxa[n], t3
taxa[n],t3tip.label)
len <- t3
e
d
g
e
.
l
e
n
g
t
h
[
m
a
t
c
h
(
n
u
m
,
t
3
edge.length[match(num, t3
edge.length[match(num,t3edge[, 2])] * (2/3)
t3 <- bind.tip(t3, tip.label = data
s
p
e
c
i
e
s
[
i
]
,
e
d
g
e
.
l
e
n
g
t
h
=
l
e
n
,
w
h
e
r
e
=
n
u
m
,
p
o
s
i
t
i
o
n
=
l
e
n
)
n
F
species[i], edge.length = len, where = num, position = len) nF
species[i],edge.length=len,where=num,position=len)nFgen.n[n] <- g + 1
nF
s
p
.
n
[
n
]
<
−
s
+
1
n
u
m
<
−
g
r
e
p
(
n
F
sp.n[n] <- s + 1 num <- grep(nF
sp.n[n]<−s+1num<−grep(nFtaxa[n], t3
t
i
p
.
l
a
b
e
l
)
t
3
tip.label) t3
tip.label)t3node.label[match(t3
e
d
g
e
[
m
a
t
c
h
(
n
u
m
,
t
3
edge[match(num, t3
edge[match(num,t3edge[, 2]), 1], unique(t3
e
d
g
e
[
,
1
]
)
)
]
<
−
p
a
s
t
e
(
"
N
N
"
,
t
3
edge[, 1]))] <- paste("NN", t3
edge[,1]))]<−paste("NN",t3Nnode + 1, sep = “”)
rnN3
n
o
d
e
.
l
a
b
e
l
[
m
a
t
c
h
(
n
F
node.label[match(nF
node.label[match(nFbn[n], rnN3
n
o
d
e
.
l
a
b
e
l
)
]
<
−
p
a
s
t
e
(
"
N
N
"
,
t
3
node.label)]<- paste("NN", t3
node.label)]<−paste("NN",t3Nnode + 1, sep = “”)
nF
b
n
[
n
]
<
−
p
a
s
t
e
(
"
N
N
"
,
t
3
bn[n] <- paste("NN", t3
bn[n]<−paste("NN",t3Nnode + 1, sep = “”)
nFKaTeX parse error: Expected 'EOF', got '}' at position 33: … }̲ …edge[, 1])[match(nF
b
n
[
n
]
,
t
3
bn[n], t3
bn[n],t3node.label)]
if ((2/3)*nF
r
n
.
b
l
[
n
]
<
=
n
F
rn.bl[n] <= nF
rn.bl[n]<=nFbn.bl[n]) { len <-(nF
r
n
.
b
l
[
n
]
−
n
F
rn.bl[n]-nF
rn.bl[n]−nFbn.bl[n])/2 }
if ((2/3)*nF
r
n
.
b
l
[
n
]
>
n
F
rn.bl[n] > nF
rn.bl[n]>nFbn.bl[n]) { len <-nF
r
n
.
b
l
[
n
]
∗
2
/
3
−
n
F
rn.bl[n]*2/3-nF
rn.bl[n]∗2/3−nFbn.bl[n] }
t3 <- bind.tip(t3, tip.label = data
s
p
e
c
i
e
s
[
i
]
,
e
d
g
e
.
l
e
n
g
t
h
=
l
e
n
,
w
h
e
r
e
=
n
u
m
,
p
o
s
i
t
i
o
n
=
l
e
n
)
n
F
species[i], edge.length = len, where = num, position = len) nF
species[i],edge.length=len,where=num,position=len)nFgen.n[n] <- g + 1
nF
s
p
.
n
[
n
]
<
−
s
+
1
n
u
m
<
−
u
n
i
q
u
e
(
t
3
sp.n[n] <- s + 1 num <- unique(t3
sp.n[n]<−s+1num<−unique(t3edge[, 1])[match(nF
b
n
[
n
]
,
t
3
bn[n], t3
bn[n],t3node.label)]
t3
n
o
d
e
.
l
a
b
e
l
[
m
a
t
c
h
(
t
3
node.label[match(t3
node.label[match(t3edge[match(num, t3
e
d
g
e
[
,
2
]
)
,
1
]
,
u
n
i
q
u
e
(
t
3
edge[, 2]), 1], unique(t3
edge[,2]),1],unique(t3edge[, 1]))] <- paste(“NN”, t3
N
n
o
d
e
+
1
,
s
e
p
=
"
"
)
r
n
N
3
Nnode + 1, sep = "") rnN3
Nnode+1,sep="")rnN3node.label[match(nF
b
n
[
n
]
,
r
n
N
3
bn[n], rnN3
bn[n],rnN3node.label)]<- paste(“NN”, t3
N
n
o
d
e
+
1
,
s
e
p
=
"
"
)
n
F
Nnode + 1, sep = "") nF
Nnode+1,sep="")nFbn[n] <- paste(“NN”, t3
N
n
o
d
e
+
1
,
s
e
p
=
"
"
)
n
F
Nnode + 1, sep = "") nF
Nnode+1,sep="")nFbn.bl[n] <- len+nFKaTeX parse error: Expected 'EOF', got '}' at position 26: … }̲ …edge[, 1])[match(nF
b
n
[
n
]
,
t
3
bn[n], t3
bn[n],t3node.label)]
len <- nF
b
n
.
b
l
[
n
]
t
3
<
−
b
i
n
d
.
t
i
p
(
t
3
,
t
i
p
.
l
a
b
e
l
=
d
a
t
a
bn.bl[n] t3 <- bind.tip(t3, tip.label = data
bn.bl[n]t3<−bind.tip(t3,tip.label=dataspecies[i], edge.length = len, where = num)
}
}
}
data <- add.tip[add.tipKaTeX parse error: Expected '}', got 'EOF' at end of input: …grep(paste(datagenus[i], "”, sep = “”), t3KaTeX parse error: Expected '}', got 'EOF' at end of input: … len <- t3edge.length[match(n, t3
e
d
g
e
[
,
2
]
)
]
/
2
t
3
<
−
b
i
n
d
.
t
i
p
(
t
3
,
t
i
p
.
l
a
b
e
l
=
d
a
t
a
edge[, 2])]/2 t3 <- bind.tip(t3, tip.label = data
edge[,2])]/2t3<−bind.tip(t3,tip.label=dataspecies[i], edge.length = len, where = n, position = len)
nG
s
p
.
n
[
m
a
t
c
h
(
d
a
t
a
sp.n[match(data
sp.n[match(datagenus[i], nG
g
e
n
u
s
)
]
<
−
l
e
n
g
t
h
(
n
)
+
1
n
u
<
−
g
r
e
p
(
p
a
s
t
e
(
d
a
t
a
genus)] <- length(n) + 1 nu <- grep(paste(data
genus)]<−length(n)+1nu<−grep(paste(datagenus[i], “_”, sep = “”), t3
t
i
p
.
l
a
b
e
l
)
n
u
m
<
−
f
a
s
t
M
R
C
A
(
t
3
,
t
3
tip.label) num <- fastMRCA(t3, t3
tip.label)num<−fastMRCA(t3,t3tip.label[nu[1]], t3
t
i
p
.
l
a
b
e
l
[
n
u
[
2
]
]
)
t
3
tip.label[nu[2]]) t3
tip.label[nu[2]])t3node.label[match(num, unique(t3
e
d
g
e
[
,
1
]
)
)
]
<
−
p
a
s
t
e
(
"
N
N
"
,
t
3
edge[, 1]))] <- paste("NN", t3
edge[,1]))]<−paste("NN",t3Nnode + 1, sep = “”)
rnN3
n
o
d
e
.
l
a
b
e
l
[
m
a
t
c
h
(
n
G
node.label[match(nG
node.label[match(nGbn[n], rnN3
n
o
d
e
.
l
a
b
e
l
)
]
<
−
p
a
s
t
e
(
"
N
N
"
,
t
3
node.label)]<- paste("NN", t3
node.label)]<−paste("NN",t3Nnode + 1, sep = “”)
nG
b
n
[
m
a
t
c
h
(
d
a
t
a
bn[match(data
bn[match(datagenus[i], nG
g
e
n
u
s
)
]
<
−
p
a
s
t
e
(
"
N
N
"
,
t
3
genus)] <- paste("NN", t3
genus)]<−paste("NN",t3Nnode + 1, sep = “”)
nG
b
n
.
b
l
[
m
a
t
c
h
(
d
a
t
a
bn.bl[match(data
bn.bl[match(datagenus[i], nGKaTeX parse error: Expected 'EOF', got '}' at position 32: … }̲ …tip.label[min(n)], t3
t
i
p
.
l
a
b
e
l
[
m
a
x
(
n
)
]
)
l
e
n
<
−
a
s
.
n
u
m
e
r
i
c
(
b
r
a
n
c
h
i
n
g
.
t
i
m
e
s
(
t
3
)
)
[
m
a
t
c
h
(
n
u
m
,
u
n
i
q
u
e
(
t
3
tip.label[max(n)]) len <- as.numeric(branching.times(t3))[match(num, unique(t3
tip.label[max(n)])len<−as.numeric(branching.times(t3))[match(num,unique(t3edge[, 1]))]
t3 <- bind.tip(t3, tip.label = dataKaTeX parse error: Expected 'EOF', got '}' at position 61: … }̲ } …tip.label), which(!is.na(match(t3
t
i
p
.
l
a
b
e
l
,
s
p
L
i
s
t
tip.label, spList
tip.label,spListspecies))))
t3 <- drop.tip(t3, tip = toDrop)
Re <- which(!is.na(match(t3
n
o
d
e
.
l
a
b
e
l
,
r
n
N
3
node.label, rnN3
node.label,rnN3node.label)))
noRe <- which(is.na(match(t3
n
o
d
e
.
l
a
b
e
l
,
r
n
N
3
node.label, rnN3
node.label,rnN3node.label)))
t3
n
o
d
e
.
l
a
b
e
l
[
R
e
]
<
−
r
n
N
3
node.label[Re] <- rnN3
node.label[Re]<−rnN3oriN[match(t3
n
o
d
e
.
l
a
b
e
l
,
r
n
N
3
node.label, rnN3
node.label,rnN3node.label)[Re]]
t3$node.label[noRe] <- “”
}
else {
t3 <- NULL
}
if (output.spList == FALSE)
spList <- NULL
phylo <- list(Scenario.1 = t1, Scenario.2 = t2, Scenario.3 = t3, Species.list = spList.original)
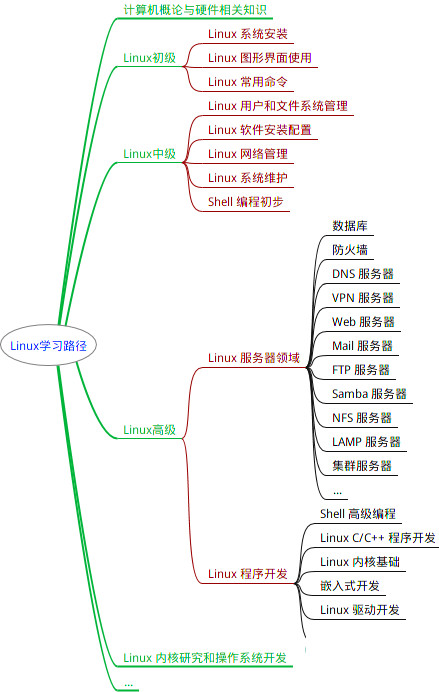
最全的Linux教程,Linux从入门到精通
======================
-
linux从入门到精通(第2版)
-
Linux系统移植
-
Linux驱动开发入门与实战
-
LINUX 系统移植 第2版
-
Linux开源网络全栈详解 从DPDK到OpenFlow

第一份《Linux从入门到精通》466页
====================
内容简介
====
本书是获得了很多读者好评的Linux经典畅销书**《Linux从入门到精通》的第2版**。本书第1版出版后曾经多次印刷,并被51CTO读书频道评为“最受读者喜爱的原创IT技术图书奖”。本书第﹖版以最新的Ubuntu 12.04为版本,循序渐进地向读者介绍了Linux 的基础应用、系统管理、网络应用、娱乐和办公、程序开发、服务器配置、系统安全等。本书附带1张光盘,内容为本书配套多媒体教学视频。另外,本书还为读者提供了大量的Linux学习资料和Ubuntu安装镜像文件,供读者免费下载。

本书适合广大Linux初中级用户、开源软件爱好者和大专院校的学生阅读,同时也非常适合准备从事Linux平台开发的各类人员。
需要《Linux入门到精通》、《linux系统移植》、《Linux驱动开发入门实战》、《Linux开源网络全栈》电子书籍及教程的工程师朋友们劳烦您转发+评论
网上学习资料一大堆,但如果学到的知识不成体系,遇到问题时只是浅尝辄止,不再深入研究,那么很难做到真正的技术提升。
一个人可以走的很快,但一群人才能走的更远!不论你是正从事IT行业的老鸟或是对IT行业感兴趣的新人,都欢迎加入我们的的圈子(技术交流、学习资源、职场吐槽、大厂内推、面试辅导),让我们一起学习成长!






















 3909
3909

 被折叠的 条评论
为什么被折叠?
被折叠的 条评论
为什么被折叠?








