# DDIMDL
DDIMDL builds multimodal deep learning framework with multiple features of drugs to predict drug-drug-interaction(DDI) events.
## Usage
*Example Usage*
```
python DDIMDL.py -f smile target enzyme -c DDIMDL -p read
```
-f *featureList*: A selection of features to be used in DDIMDL. The optional features are smile(substructure),target,enzyme and pathway of the drugs. It defaults to smile,target and enzyme.
-c *classifier*: A selection of prediction method to be used. The optional methods are DDIMDL, RF, KNN and LR. It defaults to DDIMDL.
-p *NLPProcess*: The choices are *read* and *process*. It means reading the processed result from database directly or processing the raw data again with *NLPProcess.py*. It defaults to *read*. In order to use *NLPProcess.py*, you need to install StanfordNLP package:
```
pip install stanfordnlp
```
And you need to download english package for StanforNLP:
```
import stanfordnlp
stanfordnlp.download('en')
```
## Dataset
Event.db contains the data we compiled from [DrugBank](https://www.drugbank.ca/) 5.1.3 verision. It has 4 tables:
**1.drug** contains 572 kinds of drugs and their features.
**2.event** contains the 37264 DDIs between the 572 kinds of drugs.
**3.extraction** is the process result of *NLPProcess*. Each interaction is transformed to a tuple: *{mechanism, action, drugA, drugB}*
**4.event_numer** lists the kinds of DDI events and their occurence frequency.
## Evaluation
Simply run *DDIMDL.py*, the train-test procedure will start.
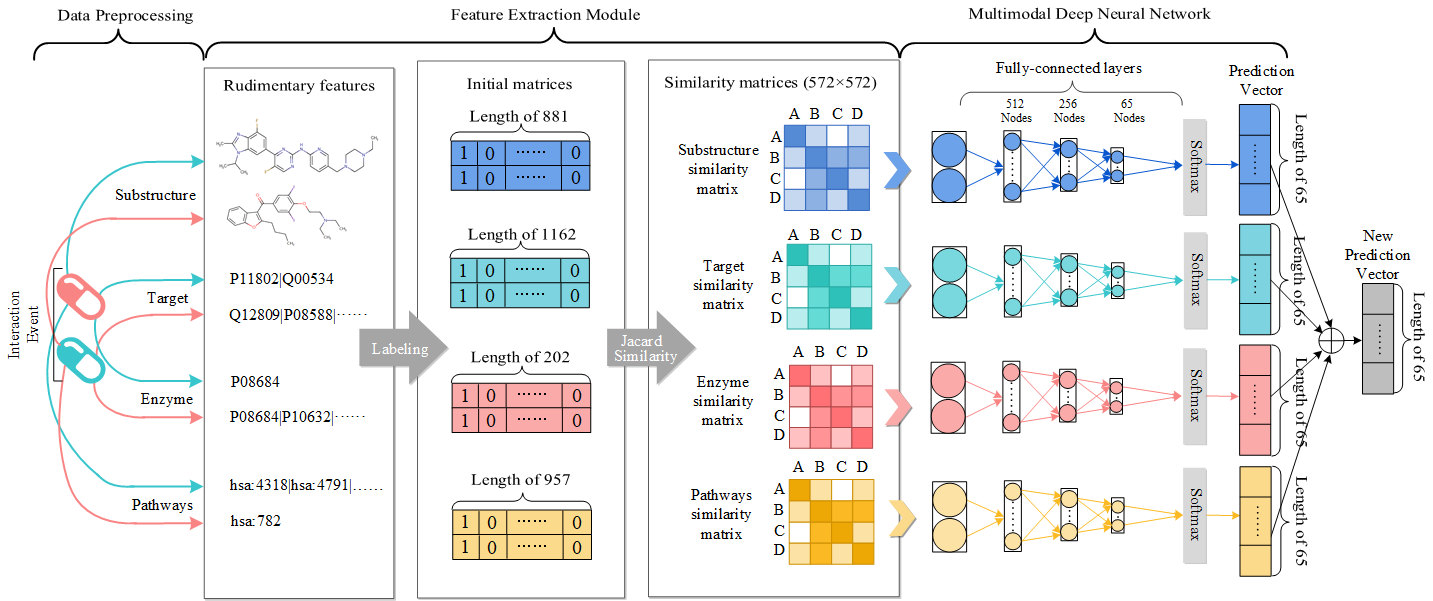
The function *prepare* will calculate the similarity between each drug based on their features.
The function *cross_validation* will take the feature matrix as input to perform 5-CV and calculate metrics. Two csv files will be generated. For example, *smile_all_DDIMDL.csv* and *smile_each_DDIMDL.csv*. The first file evaluates the method's overall performance while the other evaluates the method's performance on each event. The meaning of the metrics can be seen in array *result_all* and *result_eve* of *DDIMDL.py*.
## Requirement
- numpy (==1.18.1)
- Keras (==2.2.4)
- pandas (==1.0.1)
- scikit-learn (==0.21.2)
- stanfordnlp (==0.2.0)
Use the following command to install all dependencies.
```
pip install requirement.txt
```
**Notice: Too high version of sklearn will probably not work. We use 0.21.2 for sklearn.**
## Citation
Please kindly cite the paper if you use the code or the datasets in this repo:
```
@article{deng2020multimodal,
title={A multimodal deep learning framework for predicting drug-drug interaction events},
author={Deng, Yifan and Xu, Xinran and Qiu, Yang and Xia, Jingbo and Zhang, Wen and Liu, Shichao},
journal={Bioinformatics}
}
```





















 2062
2062











 被折叠的 条评论
为什么被折叠?
被折叠的 条评论
为什么被折叠?








