一、集成学习
1.基本定义
把多个学习器(常为弱学习器)通过一定的方式组合起来,以达到强学习器的效果。
2.常见的方法
Boosting,Bagging,Stacking,Blending。
3.常见模型融合
(1)简单加权融合
- 回归(分类概率):算术平均融合(arithmetic mean),几何平均融合(geometic mean);
- 分类:投票(voting)或者众数;
- 综合:排序融合(rank averaging),log融合。
(2)boosting/bagging
多树的提升方法,在xgboost,adaboost,GBDT
(3)stacking/blending
构建多层模型,并利用预测结果再拟合预测。
二、Boosting
1.特点
Boosting会训练一系列的弱学习器,先从初始训练集训练出一个基学习器,再根据基学习器的表现对样本分布进行调整,使得先前的基学习器做错的训练样本在后续收到更多的关注,然后基于调整后的样本分布来训练下一个基学习器;如此重复进行,直至基学习器数目达到实现指定的值T,或整个集成结果达到退出条件,最后将这些学习器加权求和。在学习过程中,后期的学习器更关注先前学习器学习中的错误。
2.主要思想
(1)adaboost思想,采用调整样本权重的方式来对样本分布进行调整,即提高前一轮个体学习器错误分类的样本的权重,而降低那些正确分类的样本的权重,这样就能使得错误分类的样本可以受到更多的关注,从而在下一轮中可以正确分类,使得分类问题被一系列的弱分类器“分而治之”。通过每次迭代改变训练数据的权值分布,使得数据在每一个基本分类器的学习中起到不同的作用,从而使数据发挥各自不同的作用,因此不易发生过拟合。
(2)Gradient Boosting思想,也是将多个弱学习器按照一定方法融合,已达到强学习器的效果,但它将损失函数梯度下降的方向作为优化的目标,新的学习器建立在之前学习器损失函数梯度下降的方向。其中代表算法有GBDT、XGBoost。
3.优缺点
(1)Boosting通常考虑同质弱学习器;
(2)个体学习器间存在强依赖关系、必须串行生成的序列化方法,故时间开销大。
三、Bagging
1.主要思想
Bagging是基于有放回重采样法。假设包含m个样本的数据集,a.随机从样本中取出一个样本放入采样集中,再把该样本放回初试数据集中(下次采样时该样本仍可能被选中);b.上一步重复m次。最终得到一个采样集D1。按照上述方法采样出T个采样集(D1,D2,...,DT),然后基于每个采样集训练出T个基学习器,再将这些基学习器进行结合以达到最终的结果。即在对测试集进行预测时,Bagging通常对分类问题采用投票法(Voting),对回归问题通常采用平均法(Averaging)。
2.优缺点
(1)Bagging通常考虑同质弱学习器,一个Bagging集成与直接使用基分类器算法训练一个学习器的复杂度同阶,说明Bagging是一个高效的集成学习算法;
(2)个体学习器之间不存在强依赖关系,相互独立地并行学习这些弱学习器;
(3)从偏差-方差的角度来说,boosting主要关注减小偏差,而Bagging主要关注降低方差,也就说明boosting在弱学习器上表现更好,而降低方差可以减小过拟合的风险,所以Bagging通常在强分类和复杂模型上表现得很好。举个例子:bagging在不减枝决策树、神经网络等易受样本扰动的学习器上效果更为明显。
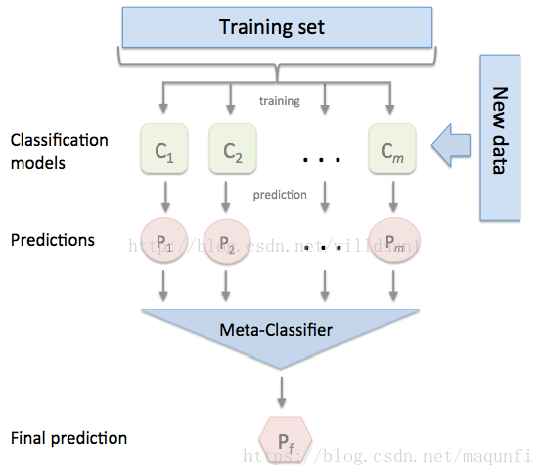
四、Stacking(堆叠法)
stacking 就是当用初始训练数据学习出若干个基学习器后,将这几个学习器的预测结果作为新的训练集,来学习一个新的学习器。接下来我将从单个基模型生成以及多个模型集成两个方面介绍。
1.单个基模型生成过程
首先将所有数据集生成测试集和训练集(假如训练集为10000,测试集为2500行),那么上层会进行5折交叉检验,使用训练集中的8000条作为训练集,剩余2000行作为验证集(橙色),下图是一个5折stacking中基模型在所有数据集上生成预测结果的过程,次学习器会基于模型的预测结果进行再训练,
每次验证相当于使用了蓝色的8000条数据训练出一个模型,使用模型对验证集进行验证得到2000条数据,并对测试集进行预测,得到2500条数据,这样经过5次交叉检验,可以得到中间的橙色的5*2000条验证集的结果(相当于每条数据的预测结果),5*2500条测试集的预测结果。
接下来会将验证集的5*2000条预测结果拼接成10000行长的矩阵,得到一个维度为10000的列向量,记为A1;对于5*2500行的测试集的预测结果进行加权平均(注意测试集加权平均),得到一个维度为2500的列向量,记为B1。 于是单个基模型可得到新的数据集:新训练集(A1, 原来训练集标签), 测试集B1。
2.多个基模型集成
假设我们有3个基模型,每个基模型都会生成新的数据集:训练集A1、A2、A3(三个特征), 测试集B1、B2、B3。新数据集维度说明,训练集:训练集样本数*基模型数,测试集:测试集样本数*基模型数。
最后用一个分类器学习新数据集,然后预测,以得到最终的结果。
原文链接:https://blog.csdn.net/maqunfi/article/details/82220115
3.代码实现
"""
Purpose: This script tries to implement a technique called stacking/blending/stacked generalization.
The reason I have to make this a runnable script because I found that there isn't really any
readable code that demonstrates this technique. You may find the pseudocode in various papers but they
are all each kind of different.
Dataset is available here : http://archive.ics.uci.edu/ml/datasets/Vertebral+Column
Author: Eric Chio "log0" <im.ckieric@gmail.com>
======================================================================================================
Summary:
Just to test an implementation of stacking. Using a cross-validated random forest and SVMs, I was
only able to achieve an accuracy of about 88% (with 1000 trees and up). Using stacked generalization
I have seen a maximum of 93.5% accuracy. It does take runs to find it out though. This uses only
(10, 20, 10) trees for the three classifiers.
This code is heavily inspired from the code shared by Emanuele (https://github.com/emanuele) , but I
have cleaned it up to makeit available for easy download and execution.
======================================================================================================
Methodology:
Three classifiers (RandomForestClassifier, ExtraTreesClassifier and a GradientBoostingClassifier
are built to be stacked by a LogisticRegression in the end.
Some terminologies first, since everyone has their own, I'll define mine to be clear:
- DEV SET, this is to be split into the training and validation data. It will be cross-validated.
- TEST SET, this is the unseen data to validate the generalization error of our final classifier. This
set will never be used to train.
======================================================================================================
Log Output:
X_test.shape = (62L, 6L)
blend_train.shape = (247L, 3L)
blend_test.shape = (62L, 3L)
Training classifier [0]
Fold [0]
Fold [1]
Fold [2]
Fold [3]
Fold [4]
Training classifier [1]
Fold [0]
Fold [1]
Fold [2]
Fold [3]
Fold [4]
Training classifier [2]
Fold [0]
Fold [1]
Fold [2]
Fold [3]
Fold [4]
Y_dev.shape = 247
Accuracy = 0.935483870968
======================================================================================================
Data Set Information:
Biomedical data set built by Dr. Henrique da Mota during a medical residence period in the Group
of Applied Research in Orthopaedics (GARO) of the Centre Médico-Chirurgical de Réadaptation des
Massues, Lyon, France. The data have been organized in two different but related classification
tasks. The first task consists in classifying patients as belonging to one out of three
categories: Normal (100 patients), Disk Hernia (60 patients) or Spondylolisthesis (150
patients). For the second task, the categories Disk Hernia and Spondylolisthesis were merged
into a single category labelled as 'abnormal'. Thus, the second task consists in classifying
patients as belonging to one out of two categories: Normal (100 patients) or Abnormal (210
patients). We provide files also for use within the WEKA environment.
Attribute Information:
Each patient is represented in the data set by six biomechanical attributes derived from the
shape and orientation of the pelvis and lumbar spine (in this order): pelvic incidence, pelvic
tilt, lumbar lordosis angle, sacral slope, pelvic radius and grade of spondylolisthesis. The
following convention is used for the class labels: DH (Disk Hernia), Spondylolisthesis (SL),
Normal (NO) and Abnormal (AB).
"""
import csv
import random
import numpy as np
import matplotlib.pyplot as plt
from sklearn.cross_validation import StratifiedKFold
from sklearn.linear_model import LogisticRegression
from sklearn.preprocessing import LabelEncoder
from sklearn import metrics
from sklearn.ensemble import RandomForestClassifier, ExtraTreesClassifier, GradientBoostingClassifier
def run(data):
X = np.array([ i[:-1] for i in data ], dtype=float)
Y = np.array([ i[-1] for i in data ])
# We need to transform the string output to numeric
label_encoder = LabelEncoder()
label_encoder.fit(Y)
Y = label_encoder.transform(Y)
# The DEV SET will be used for all training and validation purposes
# The TEST SET will never be used for training, it is the unseen set.
dev_cutoff = len(Y) * 4/5
X_dev = X[:dev_cutoff]
Y_dev = Y[:dev_cutoff]
X_test = X[dev_cutoff:]
Y_test = Y[dev_cutoff:]
n_trees = 10
n_folds = 5
# Our level 0 classifiers
clfs = [
RandomForestClassifier(n_estimators = n_trees, criterion = 'gini'),
ExtraTreesClassifier(n_estimators = n_trees * 2, criterion = 'gini'),
GradientBoostingClassifier(n_estimators = n_trees),
]
# Ready for cross validation
skf = list(StratifiedKFold(Y_dev, n_folds))
# Pre-allocate the data
blend_train = np.zeros((X_dev.shape[0], len(clfs))) # Number of training data x Number of classifiers
blend_test = np.zeros((X_test.shape[0], len(clfs))) # Number of testing data x Number of classifiers
print 'X_test.shape = %s' % (str(X_test.shape))
print 'blend_train.shape = %s' % (str(blend_train.shape))
print 'blend_test.shape = %s' % (str(blend_test.shape))
# For each classifier, we train the number of fold times (=len(skf))
for j, clf in enumerate(clfs):
print 'Training classifier [%s]' % (j)
blend_test_j = np.zeros((X_test.shape[0], len(skf))) # Number of testing data x Number of folds , we will take the mean of the predictions later
for i, (train_index, cv_index) in enumerate(skf):
print 'Fold [%s]' % (i)
# This is the training and validation set
X_train = X_dev[train_index]
Y_train = Y_dev[train_index]
X_cv = X_dev[cv_index]
Y_cv = Y_dev[cv_index]
clf.fit(X_train, Y_train)
# This output will be the basis for our blended classifier to train against,
# which is also the output of our classifiers
blend_train[cv_index, j] = clf.predict(X_cv)
blend_test_j[:, i] = clf.predict(X_test)
# Take the mean of the predictions of the cross validation set
blend_test[:, j] = blend_test_j.mean(1)
print 'Y_dev.shape = %s' % (Y_dev.shape)
# Start blending!
bclf = LogisticRegression()
bclf.fit(blend_train, Y_dev)
# Predict now
Y_test_predict = bclf.predict(blend_test)
score = metrics.accuracy_score(Y_test, Y_test_predict)
print 'Accuracy = %s' % (score)
return score
if __name__ == '__main__':
train_file = 'data/column_3C.dat'
data = [ i for i in csv.reader(file(train_file, 'rb'), delimiter=' ') ]
data = data[1:] # remove header
best_score = 0.0
# run many times to get a better result, it's not quite stable.
for i in xrange(1):
print 'Iteration [%s]' % (i)
random.shuffle(data)
score = run(data)
best_score = max(best_score, score)
print
print 'Best score = %s' % (best_score)五、Blending
1.Blending流程图
2.算法描述
(1)划分数据集。训练集大约60%~80%;验证集大约40%~20%;
(2)构建模型。
a.假设构建M个模型M1,M2,...,MM。第i个模型对训练集学习,并且对验证集以及测试集分别进行预测,得到预测结果分别为为VMi,PMi;
b.每个模型对验证集的预测结果VM1, VM2, ..., VMM,验证集标签,则构成一个新的训练集,并对该新训练集进行训练学习,得到一个新的模型M0;
c.每个模型对测试集的预测结果PM1, PM2,...,PMM构成一个新的预测集。最后通过新的模型M0,得到最终的预测结果。























 62
62











 被折叠的 条评论
为什么被折叠?
被折叠的 条评论
为什么被折叠?








