参考链接:高斯过程 Gaussian Processes 原理、可视化及代码实现
from mpl_toolkits.mplot3d import Axes3D
from scipy.optimize import minimize
import numpy as np
import matplotlib.pyplot as plt
import matplotlib.colors as colors
import sklearn.datasets
import numpy as np
import tensorflow as tf
from scipy.optimize import minimize
plt.rcParams['figure.dpi'] = 140
DEFAULT_X_RANGE = (-3.5, 3.5)
DEFAULT_Y_RANGE = (-2.5, 2.5)
DEFAULT_CMAP = colors.ListedColormap(["#377eb8", "#ff7f00"])
DEFAULT_NORM = colors.Normalize(vmin=0, vmax=1,)
DEFAULT_N_GRID = 100
def Normalize(data):
data = np.where(data < 0, -data, data)
mx = np.max(data)
mn = np.min(data)
out = (data-mn)/(mx-mn)
return out
class GPR():
def __init__(self, optimize=True):
self.is_fit = False
self.train_X, self.train_y = None, None
self.params = {"l": 0.18016438187655198, "sigma_f": 0.31796407973986973}
self.optimize = optimize
def fit(self, X, y):
# store train data
self.train_X = np.asarray(X)
self.train_y = np.asarray(y)
# hyper parameters optimization
def negative_log_likelihood_loss(params):
self.params["l"], self.params["sigma_f"] = params[0], params[1]
Kyy = self.kernel(self.train_X, self.train_X) + 1e-8 * np.eye(len(self.train_X))
return 0.5 * self.train_y.T.dot(np.linalg.inv(Kyy)).dot(self.train_y) + 0.5 * np.linalg.slogdet(Kyy)[1] + 0.5 * len(self.train_X) * np.log(2 * np.pi)
if self.optimize:
res = minimize(negative_log_likelihood_loss, [self.params["l"], self.params["sigma_f"]],
bounds=((1e-4, 1e4), (1e-4, 1e4)),
method='L-BFGS-B')
self.params["l"], self.params["sigma_f"] = res.x[0], res.x[1]
self.is_fit = True
def predict(self, X):
if not self.is_fit:
print("GPR Model not fit yet.")
return
X = np.asarray(X)
Kff = self.kernel(self.train_X, self.train_X) # (N, N)
Kyy = self.kernel(X, X) # (k, k)
Kfy = self.kernel(self.train_X, X) # (N, k)
Kff_inv = np.linalg.inv(Kff + 1e-8 * np.eye(len(self.train_X))) # (N, N)
mu = Kfy.T.dot(Kff_inv).dot(self.train_y)
cov = Kyy - Kfy.T.dot(Kff_inv).dot(Kfy)
return mu, cov
def kernel(self, x1, x2):
dist_matrix = np.sum(x1**2, 1).reshape(-1, 1) + np.sum(x2**2, 1) - 2 * np.dot(x1, x2.T)
return self.params["sigma_f"] ** 2 * np.exp(-0.5 / self.params["l"] ** 2 * dist_matrix)
def y(x, noise_sigma=0.0):
x = np.asarray(x)
y = np.cos(x) + np.random.normal(0, noise_sigma, size=x.shape)
return y.tolist()
def make_ood_data(sample_size=500, means=(2.5, -1.75), vars=(0.01, 0.01)):
return np.random.multivariate_normal(means, cov=np.diag(vars), size=sample_size)
def make_training_data(sample_size=500):
train_examples, train_labels = sklearn.datasets.make_moons(n_samples=2 * sample_size, noise=0.1)
# Adjust data position slightly.
train_examples[train_labels == 0] += [-0.1, 0.2]
train_examples[train_labels == 1] += [0.1, -0.2]
return train_examples, train_labels
def make_testing_data(x_range=DEFAULT_X_RANGE, y_range=DEFAULT_Y_RANGE, n_grid=DEFAULT_N_GRID):
# testing data (mesh grid over data space)
x = np.linspace(x_range[0], x_range[1], n_grid)
y = np.linspace(y_range[0], y_range[1], n_grid)
xv, yv = np.meshgrid(x, y)
return np.stack([xv.flatten(), yv.flatten()], axis=-1)
# Load the train, test and OOD datasets.
train_examples, train_labels= make_training_data(
sample_size=500)
test_examples = make_testing_data()
ood_examples = make_ood_data(sample_size=500)
# Visualize
pos_examples = train_examples[train_labels == 0]
neg_examples = train_examples[train_labels == 1]
# plt.figure(figsize=(7, 5.5))
# plt.scatter(pos_examples[:, 0], pos_examples[:, 1], c="#377eb8", alpha=0.5)
# plt.scatter(neg_examples[:, 0], neg_examples[:, 1], c="#ff7f00", alpha=0.5)
# plt.scatter(ood_examples[:, 0], ood_examples[:, 1], c="red", alpha=0.1)
# plt.legend(["Positive", "Negative", "Out-of-Domain"])
# plt.ylim(DEFAULT_Y_RANGE)
# plt.xlim(DEFAULT_X_RANGE)
# plt.show()
print("train_examples.shape, train_labels.shape",train_examples.shape, train_labels.shape)
def y_2d(x, noise_sigma=0.0):
x = np.asarray(x)
y = np.sin(0.5 * np.linalg.norm(x, axis=1))
y += np.random.normal(0, noise_sigma, size=y.shape)
return y
train_X = train_examples.tolist()
train_y = train_labels
print("train_X",np.array(train_X).shape)
print("train_y",train_y.shape)
test_d1 = np.arange(-3.5, 3.5, 7/30)
print("test_d1.shape",test_d1.shape)
test_d2 = np.arange(-2.5, 2.5, 5/30)
test_d1, test_d2 = np.meshgrid(test_d1, test_d2)
test_X = [[d1, d2] for d1, d2 in zip(test_d1.ravel(), test_d2.ravel())]
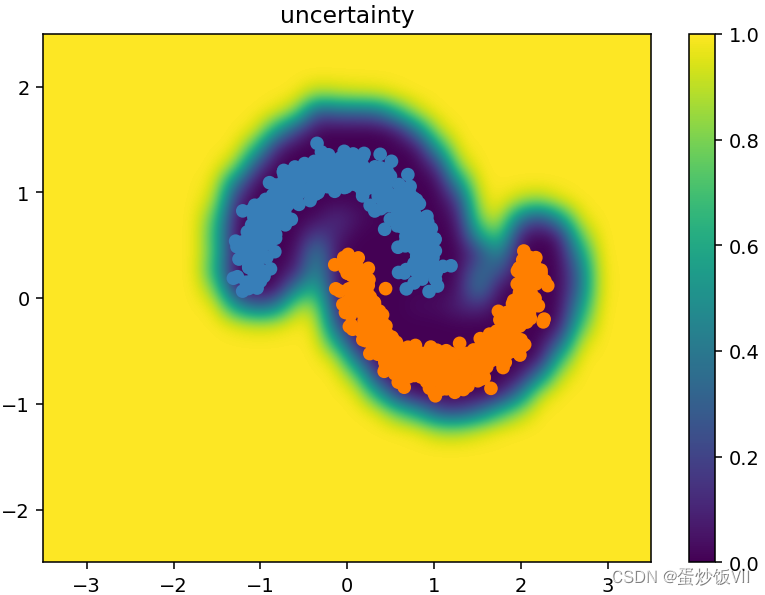
gpr = GPR(optimize=False)
gpr.fit(train_X, train_y)
mu, cov = gpr.predict(test_X)
pridict ,_= gpr.predict(train_X)
print("mu.shape",mu.shape)
print("cov.shape",cov.shape)
print("self.params",gpr.params)
uncertainty = np.diag(cov)
uncertainty = Normalize(uncertainty)
z_mu = mu.reshape(test_d1.shape)
z_uncertainty = uncertainty.reshape(test_d1.shape)
print("uncertainty.shape",uncertainty.shape)
data = np.ones(uncertainty.shape)
DEFAULT_NORM = colors.Normalize(vmin=0, vmax=1,)
fig = plt.figure(figsize=(7, 5))
plt.imshow(np.reshape(uncertainty, test_d1.shape),
origin="lower",
extent=DEFAULT_X_RANGE + DEFAULT_Y_RANGE,
vmin=DEFAULT_NORM.vmin,
vmax=DEFAULT_NORM.vmax,
interpolation='bicubic',
aspect='auto')
# plt.contourf(test_d1, test_d2, z_uncertainty, zdir='z', offset=0, alpha=0.6)
color = []
for i in pridict:
if i<0.5:
color.append("#377eb8")
else:
color.append("#ff7f00")
plt.scatter(np.asarray(train_X)[:,0], np.asarray(train_X)[:,1], c=color)
plt.colorbar()
plt.title("uncertainty")
Gpytorch实现
import os
import math
import sklearn.datasets
from matplotlib import pyplot as plt
import torch
import gpytorch
from gpytorch import kernels, means, models, mlls, settings
from gpytorch import distributions as distr
import matplotlib.colors as colors
import numpy as np
DEFAULT_X_RANGE = (-3.5, 3.5)
DEFAULT_Y_RANGE = (-2.5, 2.5)
DEFAULT_CMAP = colors.ListedColormap(["#377eb8", "#ff7f00"])
DEFAULT_NORM = colors.Normalize(vmin=0, vmax=1,)
DEFAULT_N_GRID = 100
def Normalize(data):
data = np.where(data < 0, -data, data)
m = np.mean(data)
mx = np.max(data)
mn = np.min(data)
out = (data-mn)/(mx-mn)
return out
def make_ood_data(sample_size=500, means=(2.5, -1.75), vars=(0.01, 0.01)):
return np.random.multivariate_normal(means, cov=np.diag(vars), size=sample_size)
def make_training_data(sample_size=500):
train_examples, train_labels = sklearn.datasets.make_moons(n_samples=2 * sample_size, noise=0.1)
# Adjust data position slightly.
train_examples[train_labels == 0] += [-0.1, 0.2]
train_examples[train_labels == 1] += [0.1, -0.2]
return train_examples, train_labels
def make_testing_data(x_range=DEFAULT_X_RANGE, y_range=DEFAULT_Y_RANGE, n_grid=DEFAULT_N_GRID):
# testing data (mesh grid over data space)
x = np.linspace(x_range[0], x_range[1], n_grid)
y = np.linspace(y_range[0], y_range[1], n_grid)
xv, yv = np.meshgrid(x, y)
return np.stack([xv.flatten(), yv.flatten()], axis=-1)
# Load the train, test and OOD datasets.
train_examples, train_labels= make_training_data(
sample_size=500)
test_examples = make_testing_data()
ood_examples = make_ood_data(sample_size=500)
# Visualize
pos_examples = train_examples[train_labels == 0]
neg_examples = train_examples[train_labels == 1]
# plt.figure(figsize=(7, 5.5))
# plt.scatter(pos_examples[:, 0], pos_examples[:, 1], c="#377eb8", alpha=0.5)
# plt.scatter(neg_examples[:, 0], neg_examples[:, 1], c="#ff7f00", alpha=0.5)
# plt.scatter(ood_examples[:, 0], ood_examples[:, 1], c="red", alpha=0.1)
# plt.legend(["Positive", "Negative", "Out-of-Domain"])
# plt.ylim(DEFAULT_Y_RANGE)
# plt.xlim(DEFAULT_X_RANGE)
# plt.show()
train_x = torch.from_numpy(train_examples).float()
train_y = torch.from_numpy(train_labels).float()
print("train_y",train_y.shape)
# We will use the simplest form of GP model, exact inference
class ExactGPModel(models.ExactGP):
def __init__(self, train_x, train_y, likelihood):
super(ExactGPModel, self).__init__(train_x, train_y, likelihood)
self.mean_module = means.ConstantMean()
self.covar_module = kernels.ScaleKernel(kernels.RBFKernel())
def forward(self, x):
mean_x = self.mean_module(x)
covar_x = self.covar_module(x)
return distr.MultivariateNormal(mean_x, covar_x)
# initialize likelihood and model
likelihood = gpytorch.likelihoods.GaussianLikelihood()
model = ExactGPModel(train_x, train_y, likelihood)
smoke_test = ('CI' in os.environ)
training_iter = 2 if smoke_test else 50
# Find optimal model hyperparameters
model.train()
likelihood.train()
# Use the adam optimizer
optimizer = torch.optim.Adam(model.parameters(), lr=0.1) # Includes GaussianLikelihood parameters
# "Loss" for GPs - the marginal log likelihood
mll = mlls.ExactMarginalLogLikelihood(likelihood, model)
for i in range(training_iter):
# Zero gradients from previous iteration
optimizer.zero_grad()
# Output from model
output = model(train_x)
# Calc loss and backprop gradients
loss = -mll(output, train_y)
loss.backward()
if i % 5 == 4:
print('Iter %d/%d - Loss: %.3f lengthscale: %.3f noise: %.3f' % (
i + 1, training_iter, loss.item(),
model.covar_module.base_kernel.lengthscale.item(),
model.likelihood.noise.item()
))
optimizer.step()
# Get into evaluation (predictive posterior) mode
model.eval()
likelihood.eval()
# Test points are regularly spaced along [0,1]
# Make predictions by feeding model through likelihood
test_d1 = np.arange(-3.5, 3.5, 7/30)
print("test_d1.shape",test_d1.shape)
test_d2 = np.arange(-2.5, 2.5, 5/30)
test_d1, test_d2 = np.meshgrid(test_d1, test_d2)
test_x = [[d1, d2] for d1, d2 in zip(test_d1.ravel(), test_d2.ravel())]
test_x = torch.from_numpy(np.array(test_x)).float()
with torch.no_grad(), settings.fast_pred_var():
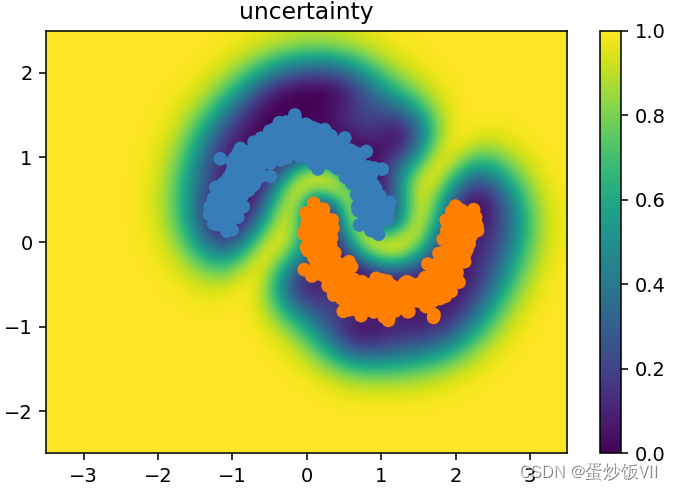
observed_pred = likelihood(model(train_x))
test_pred = likelihood(model(test_x))
color = []
print("observed_pred.mean.numpy().shape",observed_pred.mean.numpy().shape)
for i in observed_pred.mean.numpy():
if i<0.5:
color.append("#377eb8")
else:
color.append("#ff7f00")
lower, upper = test_pred.confidence_region()
uncertainty = lower.detach().numpy()+upper.detach().numpy()
uncertainty = Normalize(uncertainty)
uncertainty = uncertainty*(1-uncertainty)
uncertainty = Normalize(uncertainty)
plt.scatter(np.asarray(train_x)[:,0], np.asarray(train_x)[:,1], c=color)
plt.imshow(np.reshape(uncertainty, test_d1.shape),
origin="lower",
extent=DEFAULT_X_RANGE + DEFAULT_Y_RANGE,
vmin=DEFAULT_NORM.vmin,
vmax=DEFAULT_NORM.vmax,
interpolation='bicubic',
aspect='auto')
plt.colorbar()
plt.title("uncertainty")
稀疏高斯过程
import os
import math
import sklearn.datasets
from matplotlib import pyplot as plt
import torch
import gpytorch
from gpytorch.distributions import MultivariateNormal
from gpytorch.kernels import RBFKernel, RQKernel, MaternKernel, ScaleKernel
from gpytorch import kernels, means, models, mlls, settings
from gpytorch.means import ConstantMean
from gpytorch.models import ApproximateGP
from gpytorch.variational import (
CholeskyVariationalDistribution,
IndependentMultitaskVariationalStrategy,
VariationalStrategy,
)
import matplotlib.colors as colors
import numpy as np
from sklearn import cluster
DEFAULT_X_RANGE = (-3.5, 3.5)
DEFAULT_Y_RANGE = (-2.5, 2.5)
DEFAULT_CMAP = colors.ListedColormap(["#377eb8", "#ff7f00"])
DEFAULT_NORM = colors.Normalize(vmin=0, vmax=1,)
DEFAULT_N_GRID = 100
def Normalize(data):
data = np.where(data < 0, -data, data)
m = np.mean(data)
mx = np.max(data)
mn = np.min(data)
out = (data-mn)/(mx-mn)
return out
def make_ood_data(sample_size=500, means=(2.5, -1.75), vars=(0.01, 0.01)):
return np.random.multivariate_normal(means, cov=np.diag(vars), size=sample_size)
def make_training_data(sample_size=500):
train_examples, train_labels = sklearn.datasets.make_moons(n_samples=2 * sample_size, noise=0.1)
# Adjust data position slightly.
train_examples[train_labels == 0] += [-0.1, 0.2]
train_examples[train_labels == 1] += [0.1, -0.2]
return train_examples, train_labels
def make_testing_data(x_range=DEFAULT_X_RANGE, y_range=DEFAULT_Y_RANGE, n_grid=DEFAULT_N_GRID):
# testing data (mesh grid over data space)
x = np.linspace(x_range[0], x_range[1], n_grid)
y = np.linspace(y_range[0], y_range[1], n_grid)
xv, yv = np.meshgrid(x, y)
return np.stack([xv.flatten(), yv.flatten()], axis=-1)
# Load the train, test and OOD datasets.
train_examples, train_labels= make_training_data(
sample_size=500)
test_examples = make_testing_data()
ood_examples = make_ood_data(sample_size=500)
# Visualize
# pos_examples = train_examples[train_labels == 0]
# neg_examples = train_examples[train_labels == 1]
# plt.figure(figsize=(7, 5.5))
# plt.scatter(pos_examples[:, 0], pos_examples[:, 1], c="#377eb8", alpha=0.5)
# plt.scatter(neg_examples[:, 0], neg_examples[:, 1], c="#ff7f00", alpha=0.5)
# plt.scatter(ood_examples[:, 0], ood_examples[:, 1], c="red", alpha=0.1)
# plt.legend(["Positive", "Negative", "Out-of-Domain"])
# plt.ylim(DEFAULT_Y_RANGE)
# plt.xlim(DEFAULT_X_RANGE)
# plt.show()
def initial_values(train_data, n_inducing_points):
f_X_samples = torch.from_numpy(train_data)
initial_inducing_points = _get_initial_inducing_points(
train_data, n_inducing_points
)
initial_lengthscale = _get_initial_lengthscale(f_X_samples)
return initial_inducing_points, initial_lengthscale
def _get_initial_inducing_points(f_X_sample, n_inducing_points):
kmeans = cluster.MiniBatchKMeans(
n_clusters=n_inducing_points, batch_size=n_inducing_points * 10
)
kmeans.fit(f_X_sample)
initial_inducing_points = torch.from_numpy(kmeans.cluster_centers_)
return initial_inducing_points
def _get_initial_lengthscale(f_X_samples):
if torch.cuda.is_available():
f_X_samples = f_X_samples.cuda()
initial_lengthscale = torch.pdist(f_X_samples).mean()
return initial_lengthscale.cpu()
class GP(ApproximateGP):
def __init__(
self,
num_outputs,
initial_lengthscale,
initial_inducing_points,
kernel="RBF",
):
n_inducing_points = initial_inducing_points.shape[0]
variational_distribution = CholeskyVariationalDistribution(
n_inducing_points, batch_shape=torch.Size([])
)
variational_strategy = VariationalStrategy(
self, initial_inducing_points, variational_distribution
)
super().__init__(variational_strategy)
batch_shape = torch.Size([])
kwargs = {
"batch_shape": batch_shape,
}
if kernel == "RBF":
kernel = RBFKernel(**kwargs)
elif kernel == "Matern12":
kernel = MaternKernel(nu=1 / 2, **kwargs)
elif kernel == "Matern32":
kernel = MaternKernel(nu=3 / 2, **kwargs)
elif kernel == "Matern52":
kernel = MaternKernel(nu=5 / 2, **kwargs)
elif kernel == "RQ":
kernel = RQKernel(**kwargs)
else:
raise ValueError("Specified kernel not known.")
kernel.lengthscale = initial_lengthscale * torch.ones_like(kernel.lengthscale)
self.mean_module = ConstantMean()
self.covar_module = ScaleKernel(kernel)
def forward(self, x):
mean = self.mean_module(x)
covar = self.covar_module(x)
return MultivariateNormal(mean, covar)
likelihood = gpytorch.likelihoods.GaussianLikelihood()
initial_inducing_points, initial_lengthscale = initial_values(train_examples, 10)
initial_inducing_points = initial_inducing_points.float()
initial_lengthscale = initial_lengthscale.float()
print("initial_inducing_points",initial_inducing_points)
print("initial_lengthscale",initial_lengthscale)
model = GP(num_outputs=2,
initial_lengthscale=initial_lengthscale,
initial_inducing_points=initial_inducing_points)
smoke_test = ('CI' in os.environ)
training_iter = 2 if smoke_test else 300
# Find optimal model hyperparameters
model.train()
likelihood.train()
print(type(train_examples))
print(type(train_labels))
# train_x = torch.tensor(train_examples,dtype=torch.float)
train_x = torch.from_numpy(train_examples).float()
train_y = torch.from_numpy(train_labels).long()
print("train_x.shape",train_x.shape)
print("train_y.shape",train_y.shape)
# Use the adam optimizer
optimizer = torch.optim.Adam(model.parameters(), lr=0.1) # Includes GaussianLikelihood parameters
# "Loss" for GPs - the marginal log likelihood
# mll = mlls.ExactMarginalLogLikelihood(likelihood, model)
elbo_fn = mlls.VariationalELBO(likelihood, model, num_data=len(train_x))
loss_fn = lambda x, y: -elbo_fn(x, y)
for i in range(training_iter):
# Zero gradients from previous iteration
optimizer.zero_grad()
# Output from model
output = model(train_x)
# Calc loss and backprop gradients
loss = loss_fn(output, train_y)
# loss = -mll(output, train_y)
loss.backward()
if i % 5 == 4:
print('Iter %d/%d - Loss: %.3f lengthscale: %.3f' % (
i + 1, training_iter, loss.item(),
model.covar_module.base_kernel.lengthscale.item()
# model.likelihood.noise.item()
))
optimizer.step()
# Get into evaluation (predictive posterior) mode
model.eval()
likelihood.eval()
# Test points are regularly spaced along [0,1]
# Make predictions by feeding model through likelihood
test_d1 = np.arange(-3.5, 3.5, 7/30)
print("test_d1.shape",test_d1.shape)
test_d2 = np.arange(-2.5, 2.5, 5/30)
test_d1, test_d2 = np.meshgrid(test_d1, test_d2)
test_x = [[d1, d2] for d1, d2 in zip(test_d1.ravel(), test_d2.ravel())]
test_x = torch.from_numpy(np.array(test_x)).float()
with torch.no_grad(), settings.fast_pred_var():
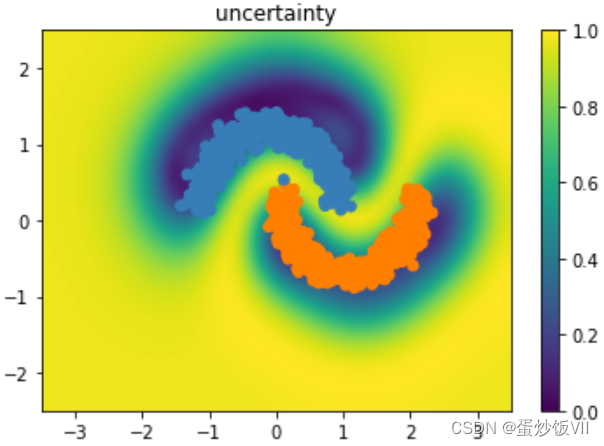
observed_pred = likelihood(model(train_x))
test_pred = likelihood(model(test_x))
color = []
print("observed_pred.mean.numpy().shape",observed_pred.mean.numpy().shape)
for i in observed_pred.mean.numpy():
if i<0.5:
color.append("#377eb8")
else:
color.append("#ff7f00")
lower, upper = test_pred.confidence_region()
uncertainty = lower.detach().numpy()+upper.detach().numpy()
uncertainty = Normalize(uncertainty)
uncertainty = uncertainty*(1-uncertainty)
uncertainty = Normalize(uncertainty)
plt.scatter(np.asarray(train_x)[:,0], np.asarray(train_x)[:,1], c=color)
plt.imshow(np.reshape(uncertainty, test_d1.shape),
origin="lower",
extent=DEFAULT_X_RANGE + DEFAULT_Y_RANGE,
vmin=DEFAULT_NORM.vmin,
vmax=DEFAULT_NORM.vmax,
interpolation='bicubic',
aspect='auto')
plt.colorbar()
plt.title("uncertainty")






















 1527
1527











 被折叠的 条评论
为什么被折叠?
被折叠的 条评论
为什么被折叠?








