基础知识点
网络分析常用指标 结果图和相关分析解释可参考如下链接文章
结果图和相关分析解释可参考如下链接文章
https://doi.org/10.1016/j.jad.2022.05.061

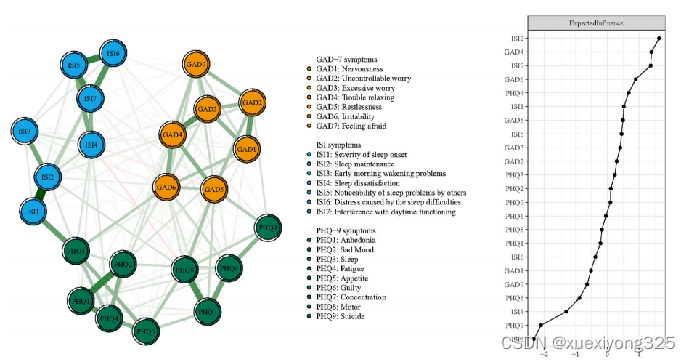
网络分析完整代码
#加载网络分析所需要的工具包
library(qgraph)
library(networktools)
library(ggplot2)
library(bootnet)
#读入数据(每行代表一个人,每列代表一个量表数据)并查看数据情况
mydata=read.csv("E:/NA/data.csv")
summary(mydata)
#根据自己的数据给每列值赋予名称
myname<-c("LT","LE","ST","SE","PMS1","PMS2","PMS3","SIS1","SIS2","SIS3","PPC1",
"PPC2","PPC3","PPC4","PSSS1","PSSS2","PSSS3","CPSS1","CPSS2")
colnames(mydata)<-myname
mydata.frame<-myname
#根据每列量表属性进行分组,比如1-4列属于Dpress(这个名字随意)
feature_group<-list(Dpress=c(1:4),PHQ=c(5:7),GAD=c(8:10),ISI=c(11:14),g4=c(15:17),g5=c(18:19))
#使用Lasso算法对相关矩阵进行稀疏化,调优参数设置为0.5
#网络的可视化采用Fruchterman-Reingold算法,连接强且数量多的节点出现在网络的中心附近,连接弱且数量少的节点行走在网络的外围
# qgraph()中使用Fruchterman-Reingold算法,布局参数需要设置为“spring”。
# R-package mgm计算predictability
# Compute node predictability 体现在网络图中每个节点外的小圆圈
library(mgm)
p <- ncol(mydata)
fit_obj <- mgm(data = mydata,
type = rep('g', p),
level = rep(1, p),
lambdaSel = 'CV',
ruleReg = 'OR',
pbar = FALSE)
pred_obj <- predict(fit_obj, mydata)
# Compute graph with tuning = 0.5 (EBIC)
# 即构建网络并使用Lasso算法对相关矩阵进行稀疏化,调优参数设置为0.5
CorMat <- cor_auto(mydata)
EBICgraph <- qgraph(CorMat, graph = "glasso", sampleSize = nrow(mydata),groups=feature_group, nodeNames=myname,
tuning = 0.5, layout = "spring", details = TRUE, pie = pred_obj$error[,2])
#该方法与上同,基本关系不变 #partial correlation计算抑郁量表相关性,但因后边需用到mynetwork,所以这步必须运行
mynetwork <- estimateNetwork(mydata, default = "EBICglasso", tuning = 0.5,corMethod="cor",corArgs=list(method="spearman",use="pairwise.complete.obs"))
plot(mynetwork,layout = "spring",nodeNames=myname,groups=feature_group,pie = pred_obj$error[,2])
#将网络图保存成pdf,EBICgraph默认跑出来的图更好看
setwd("E:/NA")
pdf("Mynetwork1.pdf")
myplot<-plot(EBICgraph,layout = "spring",nodeNames=myname,pie = pred_obj$error[,2])
dev.off()
# R-package qgraph计算expected influence
dev.new()
myplot<-plot(mynetwork,layout = "spring",nodeNames=myname,groups=feature_group)
pdf("expected influence",width=4)
c1<-centralityPlot(myplot,include = "ExpectedInfluence")
dev.off()
# R-package bootnet 检验网络的鲁棒性
b1<-bootnet(mynetwork,boots=1000,nCores=4,statistics=c("strength","expectedInfluence","edge"))
b2<-bootnet(mynetwork,boots=1000,nCores=4,type="case",statistics=c("strength","expectedInfluence","edge"))
#save bootstrapped files 保存的意义是免得下次要看结果的时候再跑,也可以选择不保存
setwd("E:/NA")
save(b1,file = "b1.Rdata")
save(b2,file = "b2.Rdata")
#下面几个就是bootstrap后的检验的一些指标,可按需
#save expectedInfluence diff test
pdf("EIDifference.pdf")
plot(b1,"expectedInfluence",order="sample",labels = TRUE)
#save edge stability graph
pdf("EdgeStability.pdf")
plot(b2,"expectedInfluence")
#Edge weights diff test
pdf("EdgeDifftest.pdf")
plot(b1,"edge",plot="difference",onlyNonZero = TRUE,order="sample")
#网络分析中第2个重要指标bridge expected influence
# R-package networktools计算bridge expected influence
library(networktools)
#constructing a partial correlation matrix
myedges<-getWmat(mynetwork)
write.csv(myedges,"MyNetcorkEdgess.csv")
#estimate bridge values for each node
#这里1代表量表组1,4个1也就是你的数据前4列是一个group
mybridge<-bridge(myplot,communities = c('1','1','1','1','2','2','2','3','3','3','4','4','4','4','5','5','5','6','6'),useCommunities="all", directed=NULL,nodes=NULL)
#下面就是画图
#create bridge graph
pdf("bridgecentrality.pdf",width=4)
plot(mybridge,include="Bridge Strength")
dev.off()
#create bridge expected influence graph
pdf("bridgeEI.pdf",width=4)
plot(mybridge,include="Bridge Expected Influence (1-step)",width=4)
dev.off()
#bridge symptoms也就是将上面计算得到的mybridge指标的第4组指标(bridge expected influence)进行排序后,取前10%或20%最大的节点,就为bridge symptoms
#sort and find bridge symptoms
a<-sort(mybridge[[4]])
#通过bootstrap来进行检验
#Bridge stability
caseDroppingBoot<-bootnet(mynetwork,boots=1000,type="case",
statistics = c("bridgeStrength","bridgeExpectedInfluence"),
communities = feature_group)
#get stability coefficients
corStability(caseDroppingBoot)
#plot centrality stability
plot(caseDroppingBoot,statistics="bridgeStrength")
plot(caseDroppingBoot,statistics="bridgeExpectedInfluence")
#Bridge stability;centraity difference
nonParametricBoot<-bootnet(mynetwork,boots=1000,type="nonparametric",
statistics = c("bridgeStrength","bridgeExpectedInfluence"),
communities = feature_group)
#plot centrality difference
plot(nonParametricBoot,statistics = "bridgeExpectedInfluence",plot="difference")
plot(nonParametricBoot,statistics = "bridgeStrength",plot="difference")
#网络对比分析,一般文章会分为男女两组人,去看网络差异
#Network Comparison test(NCT)
install.packages("NetworkComparisonTest") #下载安装工具包
library(NetworkComparisonTest)
#load data
data1<-read.table("E:/NA/gender1data.csv")
data2<-read.table("E:/NA/gender2data.csv")
#feature_group也是根据自己数据情况去设定
colnames(mydata)<-myname
mydata.frame<-myname
feature_group<-list(Dpress=c(1:10),PHQ=c(11:19),GAD=c(20:26),ISI=c(27:33))
newdata1<-na.omit(data1)
newdata2<-na.omit(data2) #NCT不能有缺失数据
#estimate networks
mynetwork1<-estimateNetwork(newdata1,default = "EBICglasso")
mynetwork2<-estimateNetwork(newdata2,default = "EBICglasso")
plot(mynetwork1,layout = "spring",nodeNames=myname,groups=feature_group)
plot(mynetwork2,layout = "spring",nodeNames=myname,groups=feature_group)
#Run NCT
myNCT1<-NCT(mynetwork1,mynetwork2,it=1000,weighted=TRUE,test.edges=FALSE,edges="ALL", test.centrality=TRUE,
centrality=c("strength","expectedInfluence"),nodes="all",)
#Get results
summary(myNCT)
至此,网络分析结束。小编也是初学者小白一个,如有任何不妥之处及任何问题都可以随时提问,欢迎指正,相互学习。这是小编写博客的初衷,利己利人,希望以后也能一如既往。
























 1万+
1万+

 被折叠的 条评论
为什么被折叠?
被折叠的 条评论
为什么被折叠?








