import numpy as np
import pandas as pd
import seaborn as sns
import matplotlib.pyplot as plt
from sklearn.datasets import load_breast_cancer
from sklearn.preprocessing import StandardScaler
from sklearn.decomposition import PCA
from mpl_toolkits import mplot3d
plt.style.use('ggplot')
cancer = load_breast_cancer()
df = pd.DataFrame(data=cancer.data,columns=cancer.feature_names)
x =df.values
print(x.shape)
scaler = StandardScaler()
scaler.fit(x)
x_scaled = scaler.transform(x)
print("x_scaled.shape====",x_scaled.shape)
# ________数据处理阶段完事————————————————————
pca_30 = PCA(n_components=30,random_state=2020)
pca_30.fit(x_scaled)
x_pac_30 =pca_30.transform(x_scaled)
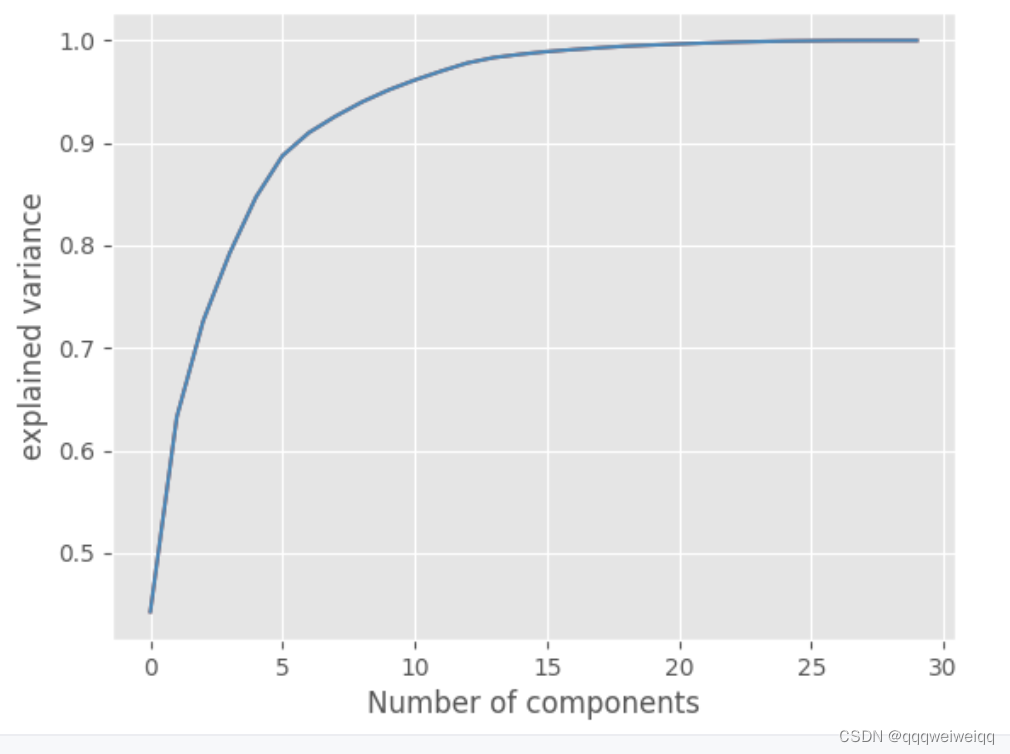
print("np.cumsum====",np.cumsum(pca_30.explained_variance_ratio_*100))
plt.plot(np.cumsum(pca_30.explained_variance_ratio_))
plt.plot(np.cumsum(pca_30.explained_variance_ratio_))
plt.xlabel("Number of components")
plt.ylabel("explained variance")
plt.savefig('elbow_plot',dpi=100)
# 3.d: Apply PCA by setting n_components=2
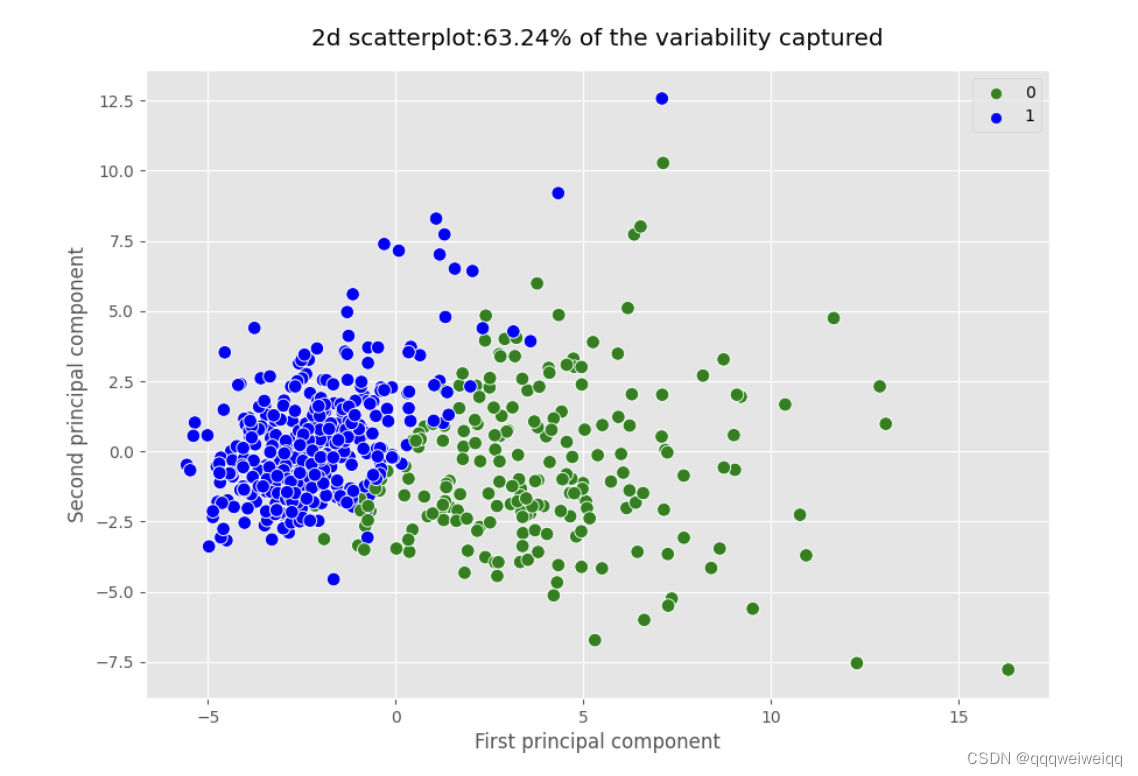
pca_2 = PCA(n_components=2,random_state=2020)
pca_2.fit(x_scaled)
x_pca_2=pca_2.transform(x_scaled)
plt.figure(figsize=(10,7))
sns.scatterplot(x=x_pca_2[:,0],y=x_pca_2[:,1],s=70,hue=cancer.target,palette=['green','blue'])
plt.title("2d scatterplot:63.24% of the variability captured",pad=15)
plt.xlabel("First principal component")
plt.ylabel("Second principal component")
plt.savefig('2d_scatterplot.png')
# ___________————————————————————————————————
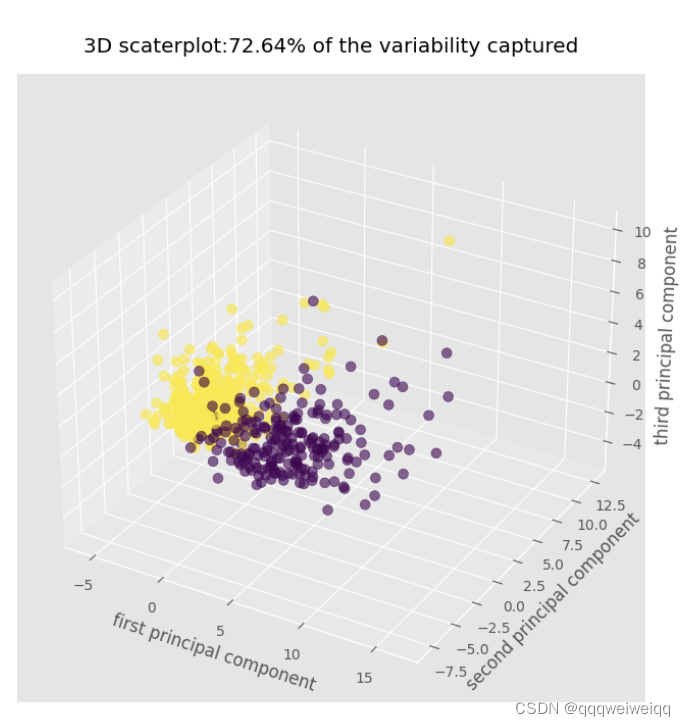
pca_3 = PCA(n_components=3,random_state=2020)
pca_3.fit(x_scaled)
x_pca_3=pca_3.transform(x_scaled)
fig= plt.figure(figsize=(12,8))
ax=plt.axes(projection='3d')
sctt=ax.scatter3D(x_pca_3[:,0],x_pca_3[:,1],x_pca_3[:,2],c=cancer.target,s=50,alpha=0.6)
plt.title("3D scaterplot:72.64% of the variability captured",pad=15)
ax.set_xlabel("first principal component")
ax.set_ylabel("second principal component")
ax.set_zlabel("third principal component")
plt.savefig('3d_scatterplot.png')
https://medium.com/data-science-365/principal-component-analysis-pca-with-scikit-learn-1e84a0c731b0







 该代码示例展示了如何使用Python的sklearn库进行主成分分析(PCA),首先对数据进行预处理,然后通过PCA降维,最后绘制2D和3D散点图以可视化主要成分,并解释了PCA在数据降维中的作用。
该代码示例展示了如何使用Python的sklearn库进行主成分分析(PCA),首先对数据进行预处理,然后通过PCA降维,最后绘制2D和3D散点图以可视化主要成分,并解释了PCA在数据降维中的作用。

















 2499
2499

 被折叠的 条评论
为什么被折叠?
被折叠的 条评论
为什么被折叠?










