存下来,实践下。
R相关的介绍:http://igraph.wikidot.com/community-detection-in-r
原文地址:http://blog.sina.com.cn/s/blog_153999aac0102vzk5.html
igraph是复杂网络分析的一个强有力的工具,纯C语言写的开源工具库,也提供了关于R和python的一些接口。里面也包含了许多模块,其中社团发现的算法就包括以下几个:
(1) edge.betweenness.community [Newman and Girvan, 2004]
(2) fastgreedy.community [Clauset et al., 2004] (modularity optimization method)
(3) label.propagation.community [Raghavan et al., 2007]
(4) leading.eigenvector.community [Newman, 2006]
(5) multilevel.community [Blondel et al., 2008] (the Louvain method)
(6) optimal.community [Brandes et al., 2008]
(7) spinglass.community [Reichardt and Bornholdt, 2006]
(8) walktrap.community [Pons and Latapy, 2005]
(9) infomap.community [Rosvall and Bergstrom, 2008]
这些算法的具体介绍请参照论文和相关资料,此次不进行介绍。
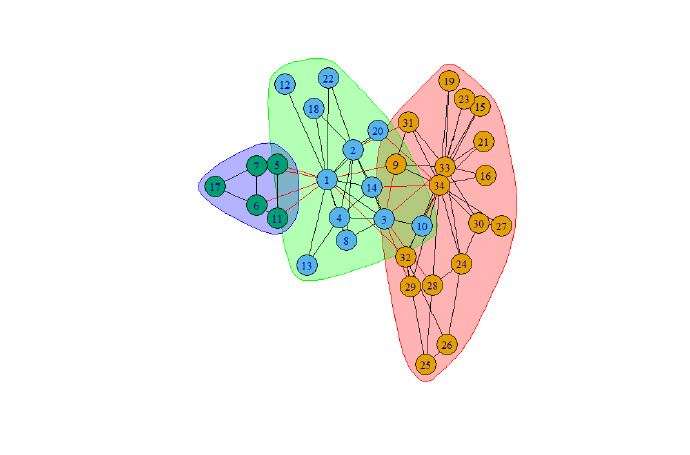
鉴于社团发现的一些重要特性,于是对该工具的如上算法进行测试,测试工具为R语言 + RStudio。测试数据为公开的karate数据。
其测试截图如下:
library(igraph)
layout(matrix(c(1,2,3,
4,5,6),nr = 2, byrow = T))
##########################################################
#经典的“Zachary 空手道俱乐部”(Zachary‟s Karate Club)社
#会网络[26]。20 世纪70 年代,Zachary 用了两年的时间观察美国一所大学的空手道
#俱乐部内部成员间关系网络。在Zachary 调查的过程中,该俱乐部的主管与校长因
#为是否提高俱乐部收费的问题产生了争执,导致该俱乐部最终分裂成了两个分别
#以主管与校长为核心的小俱乐部。就模块度优化而言,当前学者们已经广泛认为,
#该网络的最优划分是模块度Q=0.419 的4 个社团划分。
##########################################################
g <- graph.famous("Zachary")
##
#• Community structure in social and biological networks
# M. Girvan and M. E. J. Newman
#• New to version 0.6: FALSE
#• Directed edges: TRUE
#• Weighted edges: TRUE
#• Handles multiple components: TRUE
#• Runtime: |V||E|^2 ~稀疏:O(N^3)
##
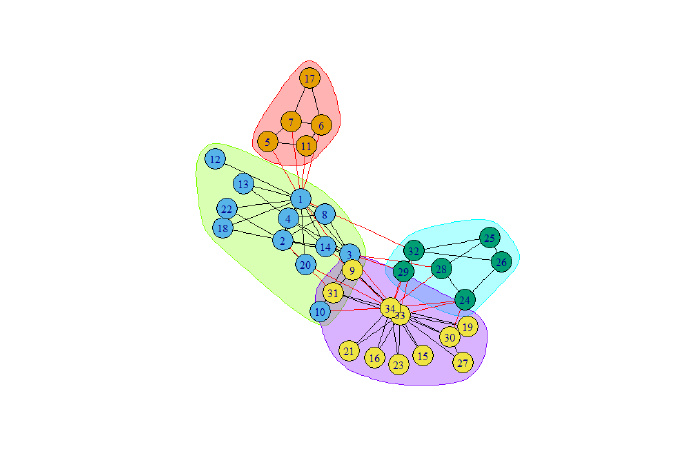
system.time(ec <- edge.betweenness.community(g))
print(modularity(ec))
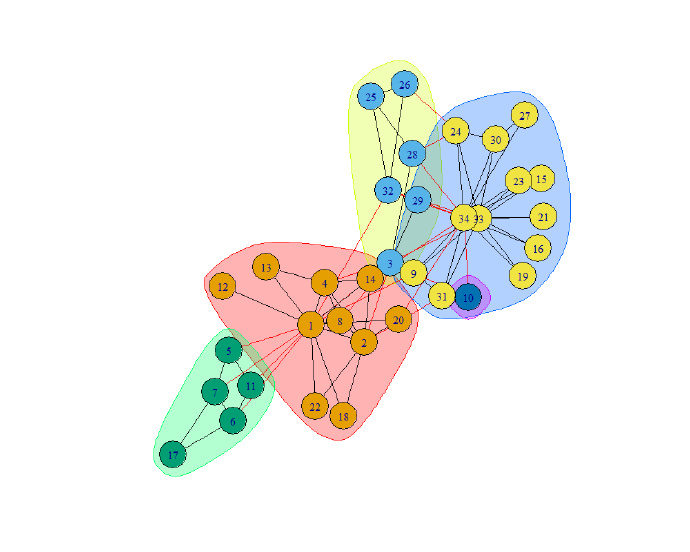
plot(ec, g)
#• Computing communities in large networks using random walks
# Pascal Pons, Matthieu Latapy
#• New to version 0.6: FALSE
#• Directed edges: FALSE
#• Weighted edges: TRUE
#• Handles multiple components: FALSE
#• Runtime: |E||V|^2
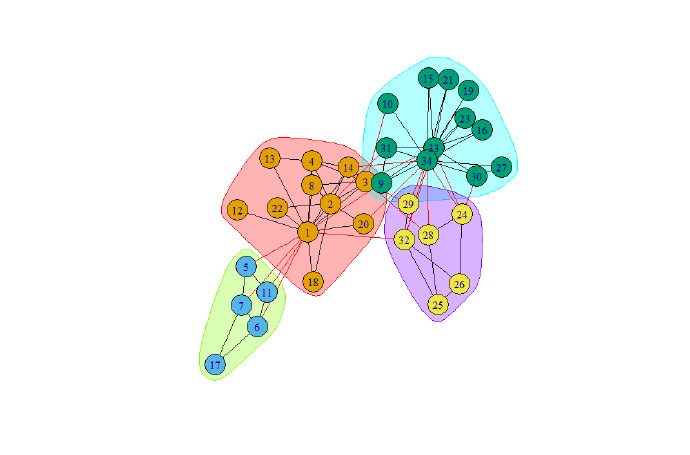
system.time(wc <- walktrap.community(g))
print(modularity(wc))
#membership(wc)
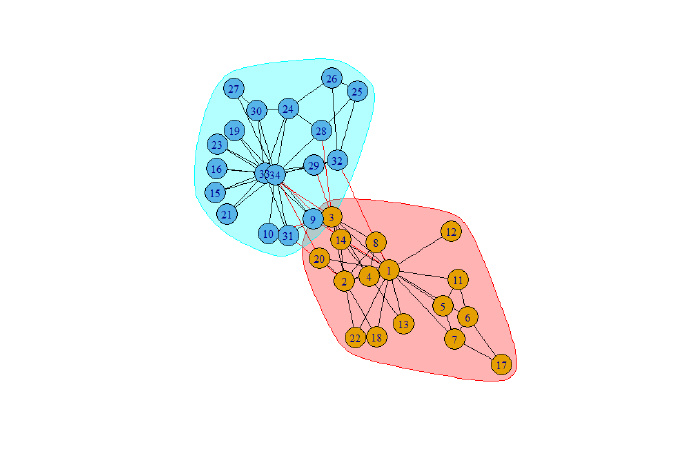
plot(wc , g)
#• Finding community structure in networks using the eigenvectors of matrices
# MEJ Newman
# Phys Rev E 74:036104 (2006)
#• New to version 0.6: FALSE
#• Directed edges: FALSE
#• Weighted edges: FALSE
#• Handles multiple components: TRUE
#• Runtime: c|V|^2 + |E| ~N(N^2)
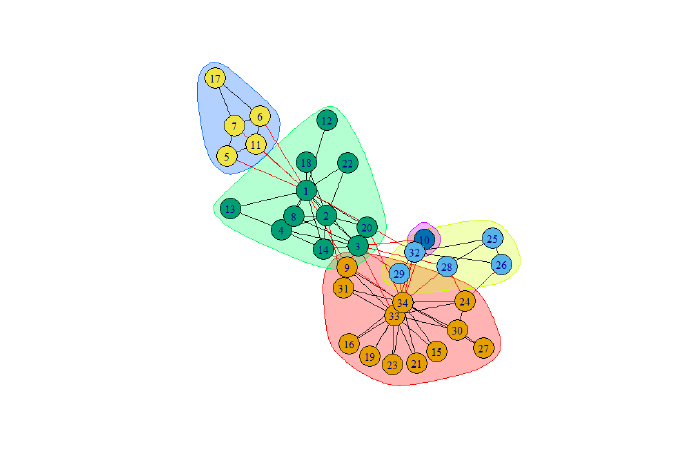
system.time(lec <-leading.eigenvector.community(g))
print(modularity(lec))
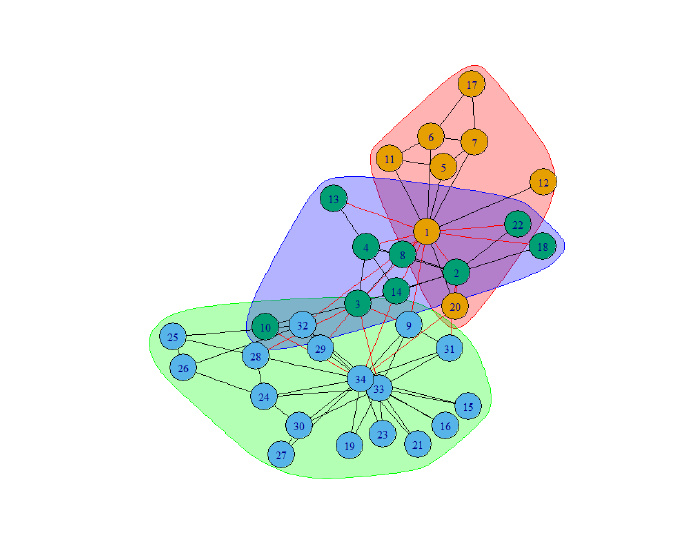
plot(lec,g)
#• Finding community structure in very large networks
# Aaron Clauset, M. E. J. Newman, Cristopher Moore
#• Finding Community Structure in Mega-scale Social Networks
# Ken Wakita, Toshiyuki Tsurumi
#• New to version 0.6: FALSE
#• Directed edges: FALSE
#• Weighted edges: TRUE
#• Handles multiple components: TRUE
#• Runtime: |V||E| log |V|
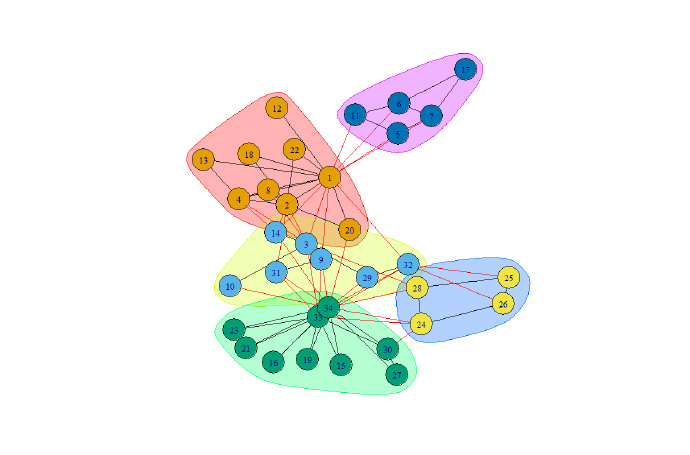
system.time(fc <- fastgreedy.community(g))
print(modularity(fc))
plot(fc, g)
#• Fast unfolding of communities in large networks
# Vincent D. Blondel, Jean-Loup Guillaume, Renaud Lambiotte, Etienne Lefebvre
#• New to version 0.6: TRUE
#• Directed edges: FALSE
#• Weighted edges: TRUE
#• Handles multiple components: TRUE
# Runtime: “linear” when |V| \approx |E| ~ sparse; (a quick glance at the algorithm \
# suggests this would be quadratic for fully-connected graphs)
system.time(mc <- multilevel.community(g, weights=NA))
print(modularity(mc))
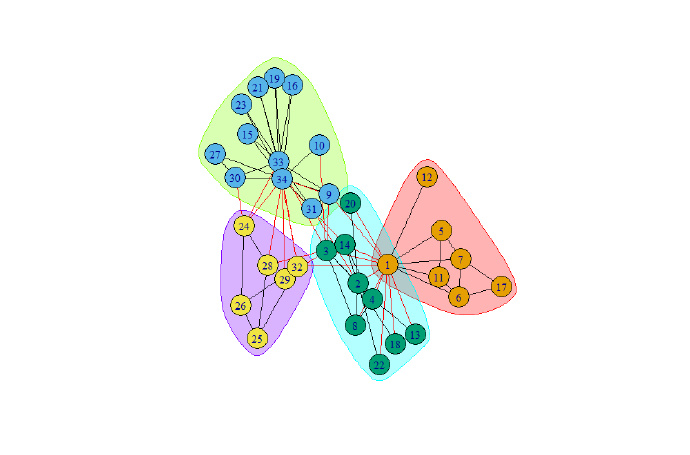
plot(mc, g)
#• Near linear time algorithm to detect community structures in large-scale networks.
# Raghavan, U.N. and Albert, R. and Kumara, S.
# Phys Rev E 76, 036106. (2007)
#• New to version 0.6: TRUE
#• Directed edges: FALSE
#• Weighted edges: TRUE
#• Handles multiple components: FALSE
# Runtime: |V| + |E|
system.time(lc <- label.propagation.community(g))
print(modularity(lc))
plot(lc , g)






















 4622
4622

 被折叠的 条评论
为什么被折叠?
被折叠的 条评论
为什么被折叠?








