

公众号“生信小课堂”
TCGA数据分析课程:生物信息学教学
相关性分析是我们生信分析中必不可少的技能,单基因的批量相关性分析,可以用于做单基因的GO,KEGG富集分析和GSEA分析,也有多基因之间的相关性分析,或者多个基因集之间的相关性分析。今天我们利用TCGA肝癌数据,挑选一些基因做相关性分析,并用不同的R包展示。
load(file = "mRNAdata.Rda")#加载数据
library(tidyverse)
library(corrplot)
library(circlize)
mRNAdata<-t(mRNAdata)#倒置数据
mRNAdata<-as.data.frame(mRNAdata)#转变为数据框
按照我们学习R语言的基础知识 R学习:R for Data Science(四)
选择列,我们随便选一些基因
new<-select(mRNAdata,LDHA,PKM,HK1,HK2,PFKFB3,SLC2A1,ATF7IP2,ZNF554)#选择列
new<-log(new+1,2)#count值取log2(count+1)计算相关性系数
cor_new<-cor(new)#计算相关性系数
cor_new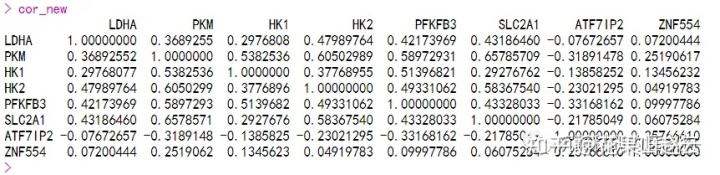
corrplot包画图
method:指定形状,可以是circle圆形(默认),square方形,ellipse,椭圆形,number数值,shade阴影,color颜色,pie饼图。type:指定显示范围,full完全(默认),lower下三角,upper上三角
corrplot(cor_new, method = "circle")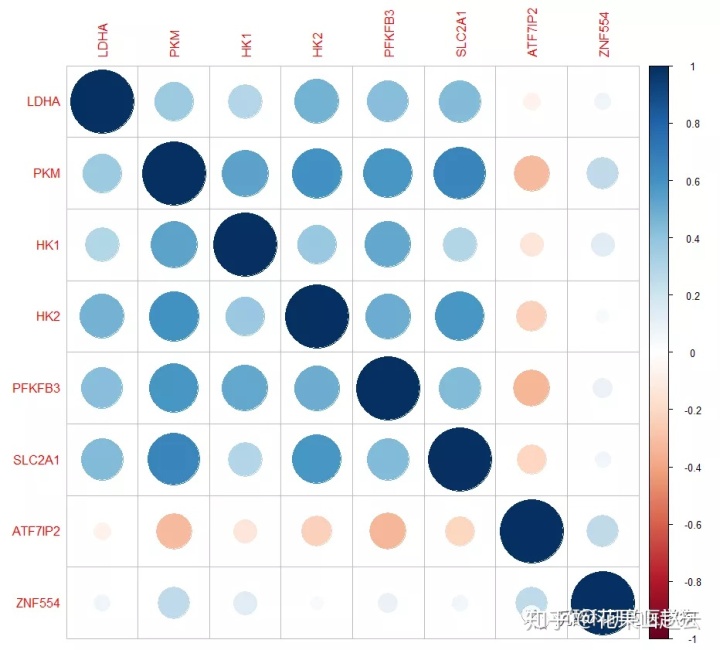
corrplot(cor_new, method = "square")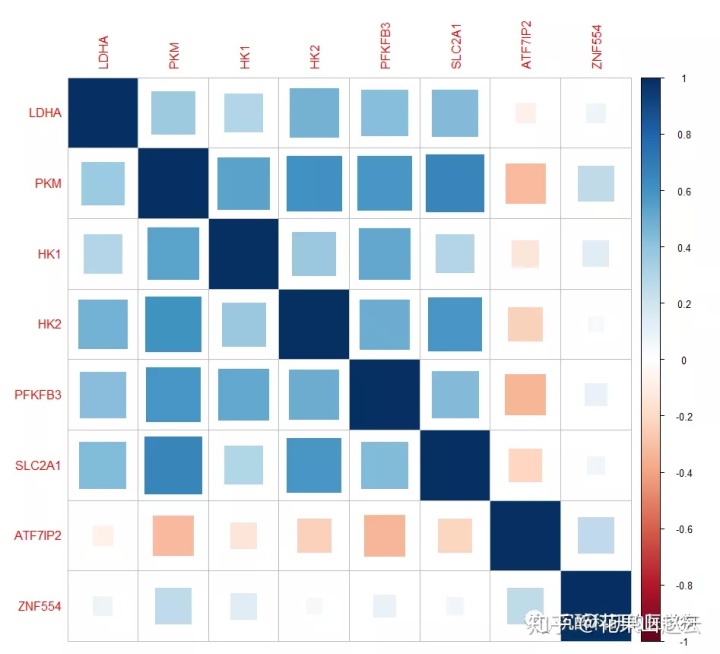
corrplot(cor_new, method = "pie")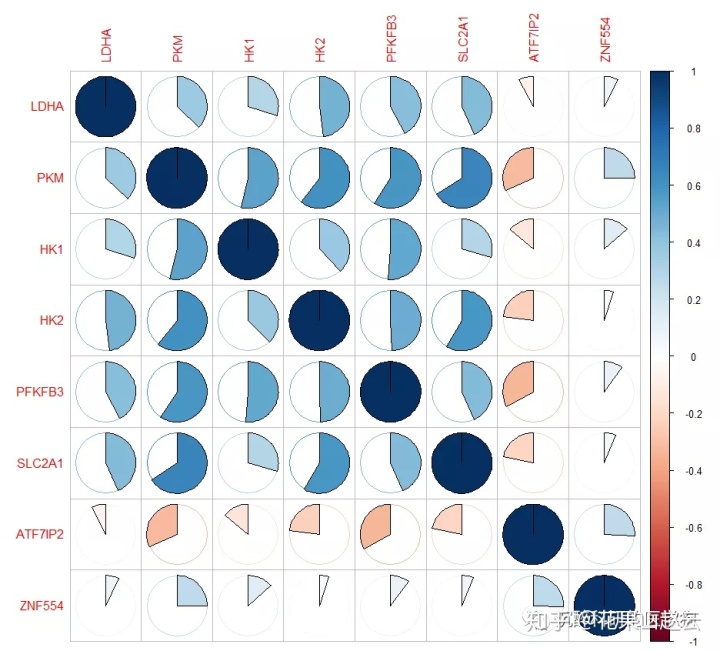
corrplot(cor_new, method = "color")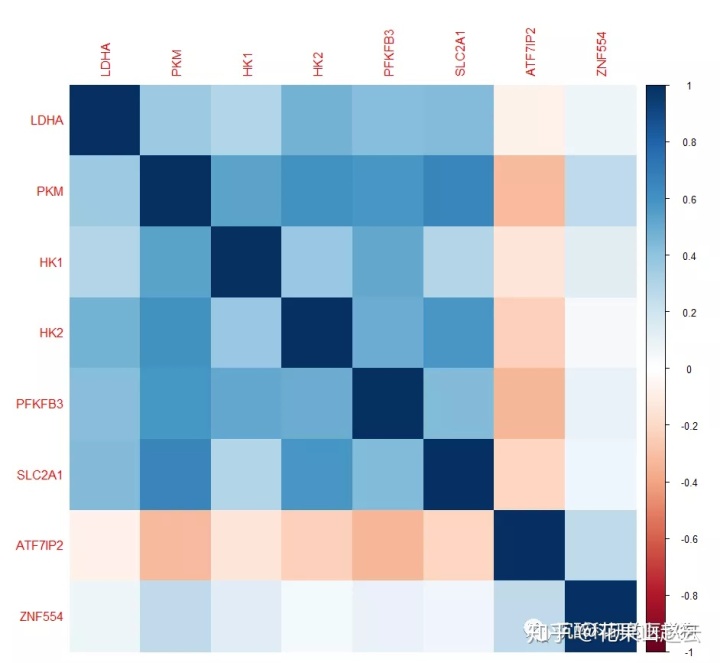
corrplot(cor_new, method = "number")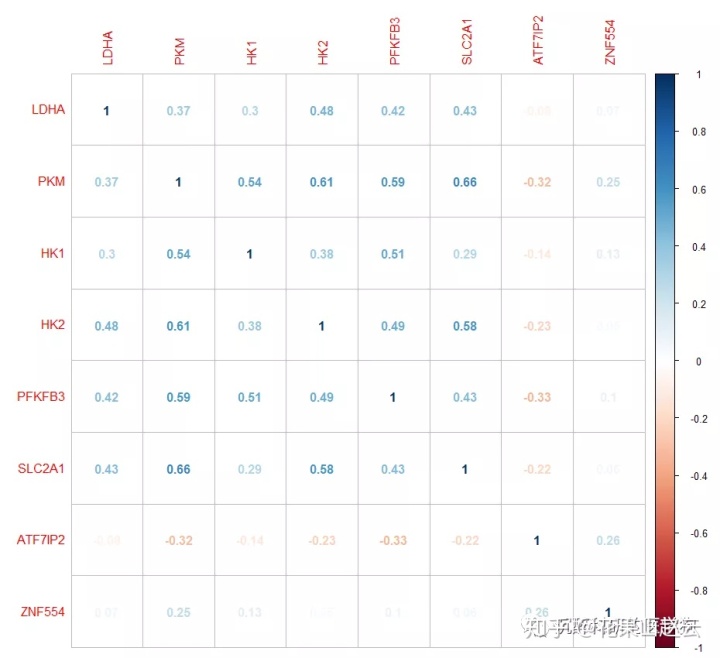
更改颜色
corrplot(cor_new, method = "color", col = colorRampPalette(c("blue", "#CD2626"))(10), title = "更改颜色")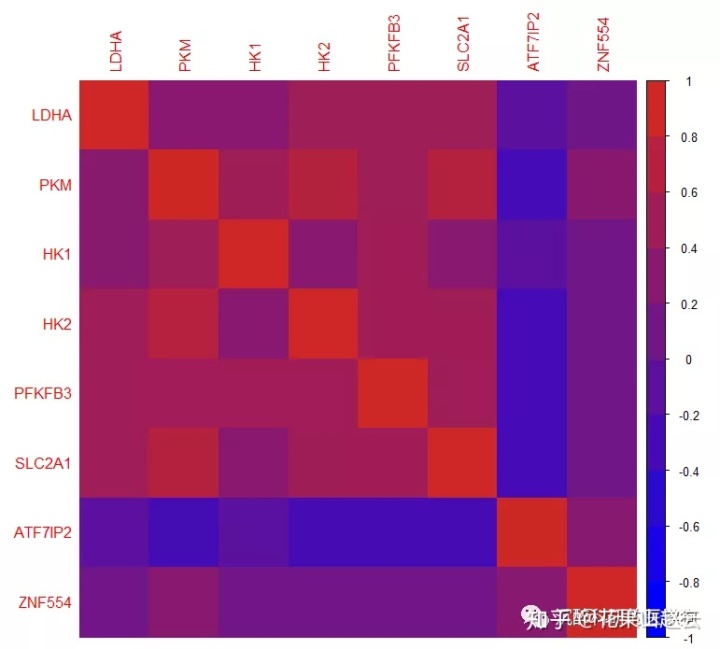
corrplot(cor_new, method = "number", col = RColorBrewer::brewer.pal(n=8, name = "RdYlGn"))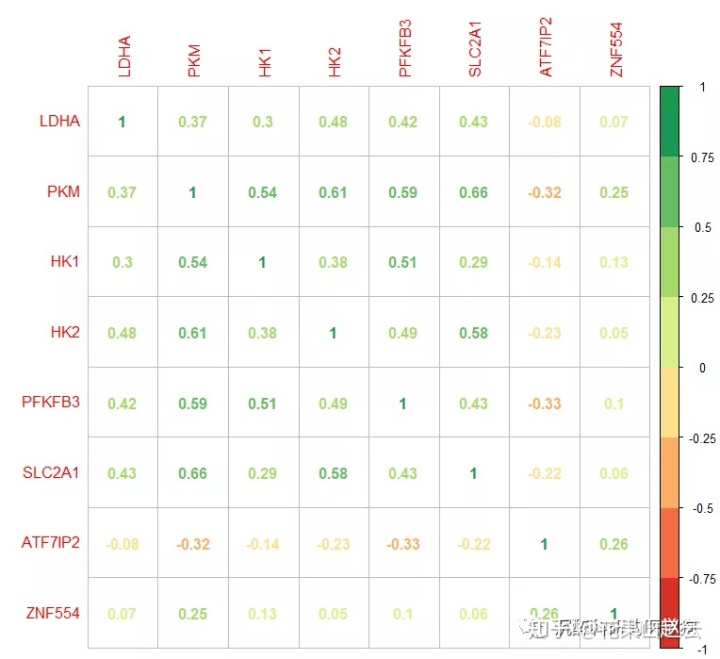
聚类显示
corrplot(cor_new,method="color",order="hclust",title = "hclust聚类", diag = TRUE,hclust.method="average",addCoef.col = "blue")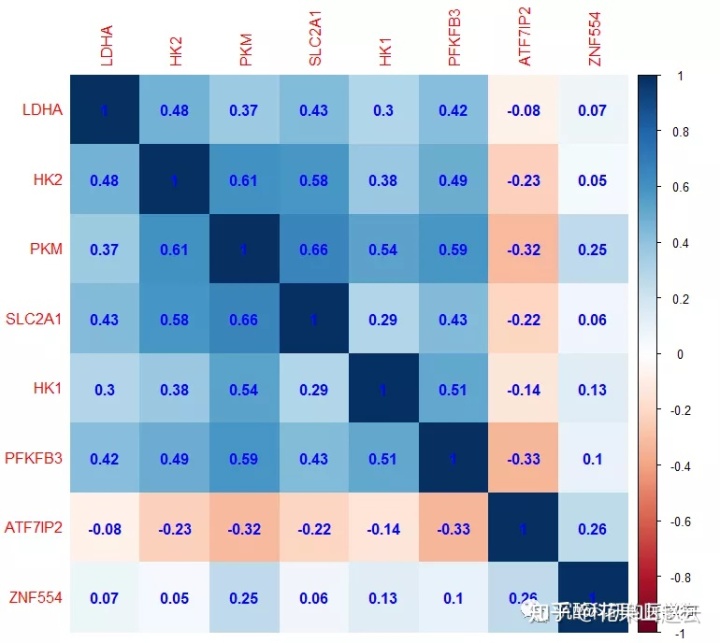
组合展示
corrplot(cor_new, method = "circle", type = "upper", tl.pos = "d")
corrplot(cor_new, add = TRUE, type = "lower", method = "number", diag = FALSE, tl.pos = "n", cl.pos = "n")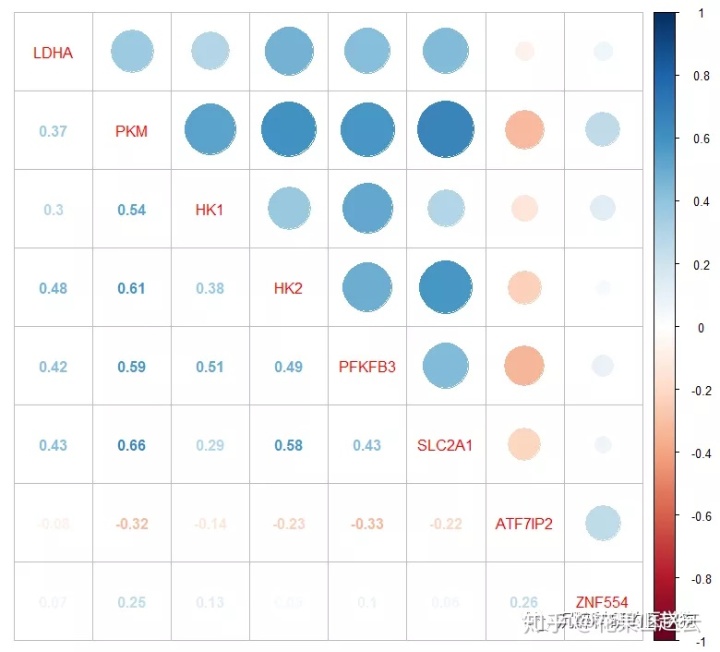
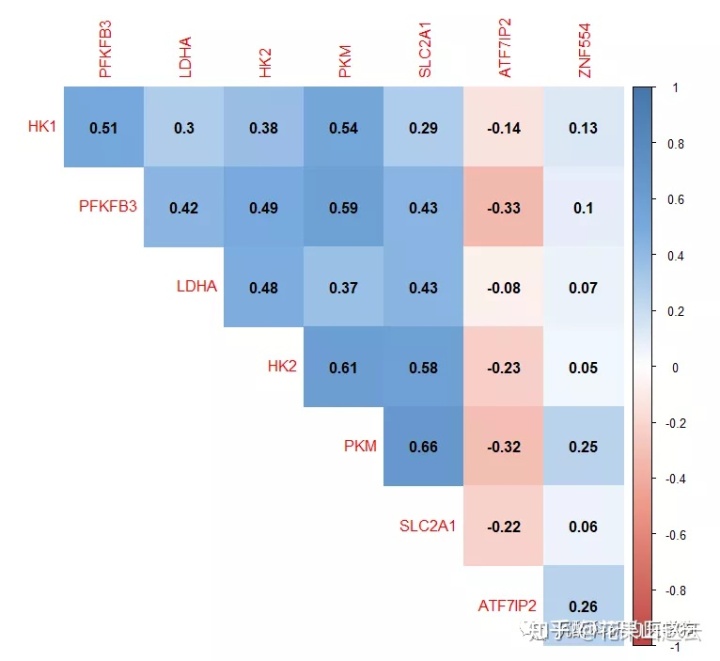
做一个好看点的
col <- colorRampPalette(c("#BB4444", "#EE9988", "#FFFFFF", "#77AADD", "#4477AA"))
corrplot(cor_new, method="color", col=col(200),
type="upper", order="hclust",
addCoef.col = "black", #添加相关系数
diag=FALSE
)
用circlize包展示
col_fun = colorRamp2(c(-1, 0, 1), c("#67BE54", "#FFFFFF", "#F82C2B"))
chordDiagram(cor_new, grid.col = 1:8, symmetric = TRUE, col = col_fun)
circos.clear()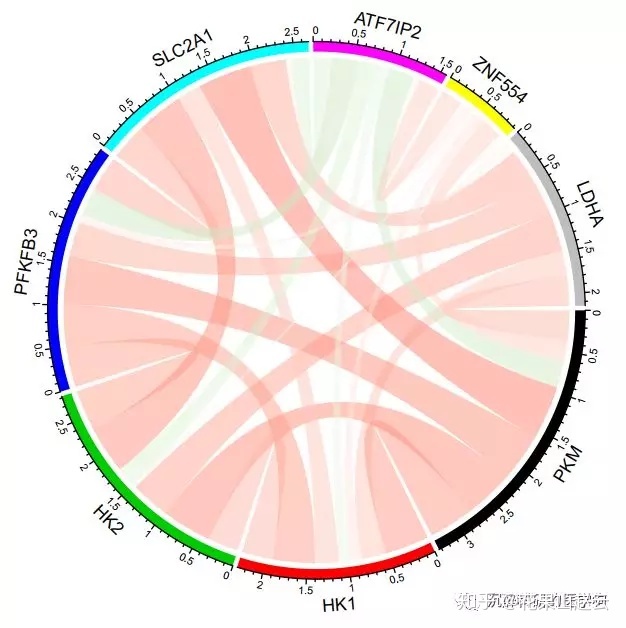
circlize包作图非常好看,我们可以用它来做一个太极
#太极图
library(circlize)
factors = 1:8
circos.par(start.degree = 22.5, gap.degree = 6)
circos.initialize(factors = factors, xlim = c(0, 1))
# yang yao is __ (a long segment)
add_yang_yao = function() {
circos.rect(0,0,1,1, col = "black")
}
# yin yao is -- (two short segments)
add_yin_yao = function() {
circos.rect(0,0,0.45,1, col = "black")
circos.rect(0.55,0,1,1, col = "black")
}
circos.track(ylim = c(0, 1), factors = factors, bg.border = NA,
panel.fun = function(x, y) {
i = get.cell.meta.data("sector.numeric.index")
if(i %in% c(2, 5, 7, 8)) add_yang_yao() else add_yin_yao()
}, track.height = 0.1)
circos.track(ylim = c(0, 1), factors = factors, bg.border = NA,
panel.fun = function(x, y) {
i = get.cell.meta.data("sector.numeric.index")
if(i %in% c(1, 6, 7, 8)) add_yang_yao() else add_yin_yao()
}, track.height = 0.1)
circos.track(ylim = c(0, 1), factors = factors, bg.border = NA,
panel.fun = function(x, y) {
i = get.cell.meta.data("sector.numeric.index")
if(i %in% c(4, 5, 6, 7)) add_yang_yao() else add_yin_yao()
}, track.height = 0.1)
# the bottom of the most recent track
r = get.cell.meta.data("cell.bottom.radius") - 0.1
# draw taiji, note default order is clock wise for `draw.sector`
draw.sector(center = c(0, 0), start.degree = 90, end.degree = -90,
rou1 = r, col = "black", border = "black")
draw.sector(center = c(0, 0), start.degree = 270, end.degree = 90,
rou1 = r, col = "white", border = "black")
draw.sector(center = c(0, r/2), start.degree = 0, end.degree = 360,
rou1 = r/2, col = "white", border = "white")
draw.sector(center = c(0, -r/2), start.degree = 0, end.degree = 360,
rou1 = r/2, col = "black", border = "black")
draw.sector(center = c(0, r/2), start.degree = 0, end.degree = 360,
rou1 = r/8, col = "black", border = "black")
draw.sector(center = c(0, -r/2), start.degree = 0, end.degree = 360,
rou1 = r/8, col = "white", border = "white")
circos.clear()
飞镖盘
factors = 1:20 # just indicate there are 20 sectors
circos.par(gap.degree = 0, cell.padding = c(0, 0, 0, 0),
start.degree = 360/20/2, track.margin = c(0, 0), clock.wise = FALSE)
circos.initialize(factors = factors, xlim = c(0, 1))
circos.track(ylim = c(0, 1), factors = factors, bg.col = "black", track.height = 0.15)
circos.trackText(x = rep(0.5, 20), y = rep(0.5, 20),
labels = c(13, 4, 18, 1, 20, 5, 12, 9, 14, 11, 8, 16, 7, 19, 3, 17, 2, 15, 10, 6),
cex = 0.8, factors = factors, col = "#EEEEEE", font = 2, facing = "downward")
circos.track(ylim = c(0, 1), factors = factors,
bg.col = rep(c("#E41A1C", "#4DAF4A"), 10), bg.border = "#EEEEEE", track.height = 0.05)
circos.track(ylim = c(0, 1), factors = factors,
bg.col = rep(c("black", "white"), 10), bg.border = "#EEEEEE", track.height = 0.275)
circos.track(ylim = c(0, 1), factors = factors,
bg.col = rep(c("#E41A1C", "#4DAF4A"), 10), bg.border = "#EEEEEE", track.height = 0.05)
circos.track(ylim = c(0, 1), factors = factors,
bg.col = rep(c("black", "white"), 10), bg.border = "#EEEEEE", track.height = 0.375)
draw.sector(center = c(0, 0), start.degree = 0, end.degree = 360,
rou1 = 0.1, col = "#4DAF4A", border = "#EEEEEE")
draw.sector(center = c(0, 0), start.degree = 0, end.degree = 360,
rou1 = 0.05, col = "#E41A1C", border = "#EEEEEE")
circos.clear()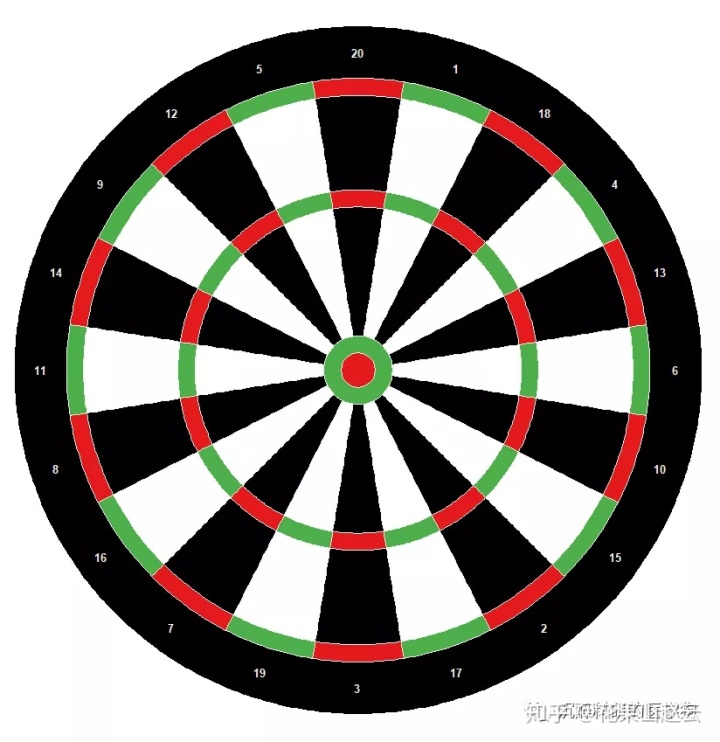
好了,多基因的相关性分析和作图就分享到这
公众号“生信小课堂”
TCGA数据分析课程:生物信息学教学







 本文通过TCGA肝癌数据,介绍如何使用R语言进行基因的相关性分析,并利用corrplot和circlize包绘制相关性图,包括圆形、方形、聚类和太极图等展示方式,为生信分析提供实用技巧。
本文通过TCGA肝癌数据,介绍如何使用R语言进行基因的相关性分析,并利用corrplot和circlize包绘制相关性图,包括圆形、方形、聚类和太极图等展示方式,为生信分析提供实用技巧。
















 4261
4261

 被折叠的 条评论
为什么被折叠?
被折叠的 条评论
为什么被折叠?








