Gradient Descent for Logistic Regression
导入所需的库
import copy, math
import numpy as np
%matplotlib widget
import matplotlib.pyplot as plt
from lab_utils_common import dlc, plot_data, plt_tumor_data, sigmoid, compute_cost_logistic
from plt_quad_logistic import plt_quad_logistic, plt_prob
plt.style.use('./deeplearning.mplstyle')
1. 数据集(多变量)
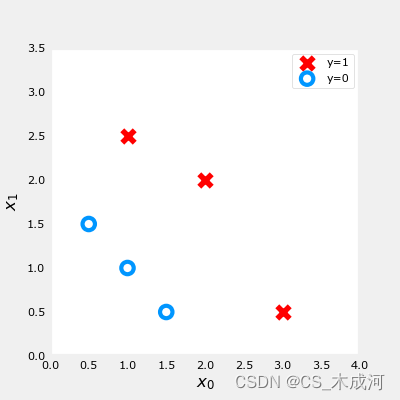
X_train = np.array([[0.5, 1.5], [1,1], [1.5, 0.5], [3, 0.5], [2, 2], [1, 2.5]])
y_train = np.array([0, 0, 0, 1, 1, 1])
fig,ax = plt.subplots(1,1,figsize=(4,4))
plot_data(X_train, y_train, ax)
ax.axis([0, 4, 0, 3.5])
ax.set_ylabel('$x_1$', fontsize=12)
ax.set_xlabel('$x_0$', fontsize=12)
plt.show()
2. 逻辑梯度下降
梯度下降计算公式:
repeat until convergence:
{
w
j
=
w
j
−
α
∂
J
(
w
,
b
)
∂
w
j
for j := 0..n-1
b
=
b
−
α
∂
J
(
w
,
b
)
∂
b
}
\begin{align*} &\text{repeat until convergence:} \; \lbrace \\ & \; \; \;w_j = w_j - \alpha \frac{\partial J(\mathbf{w},b)}{\partial w_j} \tag{1} \; & \text{for j := 0..n-1} \\ & \; \; \; \; \;b = b - \alpha \frac{\partial J(\mathbf{w},b)}{\partial b} \\ &\rbrace \end{align*}
repeat until convergence:{wj=wj−α∂wj∂J(w,b)b=b−α∂b∂J(w,b)}for j := 0..n-1(1)
其中,对于所有的
j
j
j 每次迭代同时更新
w
j
w_j
wj ,
∂
J
(
w
,
b
)
∂
w
j
=
1
m
∑
i
=
0
m
−
1
(
f
w
,
b
(
x
(
i
)
)
−
y
(
i
)
)
x
j
(
i
)
∂
J
(
w
,
b
)
∂
b
=
1
m
∑
i
=
0
m
−
1
(
f
w
,
b
(
x
(
i
)
)
−
y
(
i
)
)
\begin{align*} \frac{\partial J(\mathbf{w},b)}{\partial w_j} &= \frac{1}{m} \sum\limits_{i = 0}^{m-1} (f_{\mathbf{w},b}(\mathbf{x}^{(i)}) - y^{(i)})x_{j}^{(i)} \tag{2} \\ \frac{\partial J(\mathbf{w},b)}{\partial b} &= \frac{1}{m} \sum\limits_{i = 0}^{m-1} (f_{\mathbf{w},b}(\mathbf{x}^{(i)}) - y^{(i)}) \tag{3} \end{align*}
∂wj∂J(w,b)∂b∂J(w,b)=m1i=0∑m−1(fw,b(x(i))−y(i))xj(i)=m1i=0∑m−1(fw,b(x(i))−y(i))(2)(3)
- m 是训练集样例的数量
- f w , b ( x ( i ) ) f_{\mathbf{w},b}(x^{(i)}) fw,b(x(i)) 是模型预测值, y ( i ) y^{(i)} y(i) 是目标值
- 对于逻辑回归模型
z = w ⋅ x + b z = \mathbf{w} \cdot \mathbf{x} + b z=w⋅x+b
f w , b ( x ) = g ( z ) f_{\mathbf{w},b}(x) = g(z) fw,b(x)=g(z)
其中 g ( z ) g(z) g(z) 是 sigmoid 函数: g ( z ) = 1 1 + e − z g(z) = \frac{1}{1+e^{-z}} g(z)=1+e−z1
3. 梯度下降的实现及代码描述
实现梯度下降算法需要两步:
- 循环实现上面等式(1). 即下面的
gradient_descent - 当前梯度的计算等式(2, 3). 即下面的
compute_gradient_logistic
3.1 计算梯度
对于所有的 w j w_j wj 和 b b b,实现等式 (2),(3)
-
初始化变量计算
dj_dw和dj_db -
对每个样例:
- 计算误差 g ( w ⋅ x ( i ) + b ) − y ( i ) g(\mathbf{w} \cdot \mathbf{x}^{(i)} + b) - \mathbf{y}^{(i)} g(w⋅x(i)+b)−y(i)
- 对于这个样例中的每个输入值
x
j
(
i
)
x_{j}^{(i)}
xj(i) ,
- 误差乘以输入值
x
j
(
i
)
x_{j}^{(i)}
xj(i), 然后加到对应的
dj_dw中. (上述等式2)
- 误差乘以输入值
x
j
(
i
)
x_{j}^{(i)}
xj(i), 然后加到对应的
- 累加误差到
dj_db(上述等式3)
-
dj_db和dj_dw都除以样例总数 m m m -
在Numpy中 x ( i ) \mathbf{x}^{(i)} x(i) 是
X[i,:]或者X[i], x j ( i ) x_{j}^{(i)} xj(i) 是X[i,j]
代码描述:
def compute_gradient_logistic(X, y, w, b):
"""
Computes the gradient for linear regression
Args:
X (ndarray (m,n): Data, m examples with n features
y (ndarray (m,)): target values
w (ndarray (n,)): model parameters
b (scalar) : model parameter
Returns
dj_dw (ndarray (n,)): The gradient of the cost w.r.t. the parameters w.
dj_db (scalar) : The gradient of the cost w.r.t. the parameter b.
"""
m,n = X.shape
dj_dw = np.zeros((n,)) #(n,)
dj_db = 0.
for i in range(m):
f_wb_i = sigmoid(np.dot(X[i],w) + b) #(n,)(n,)=scalar
err_i = f_wb_i - y[i] #scalar
for j in range(n):
dj_dw[j] = dj_dw[j] + err_i * X[i,j] #scalar
dj_db = dj_db + err_i
dj_dw = dj_dw/m #(n,)
dj_db = dj_db/m #scalar
return dj_db, dj_dw
测试一下
X_tmp = np.array([[0.5, 1.5], [1,1], [1.5, 0.5], [3, 0.5], [2, 2], [1, 2.5]])
y_tmp = np.array([0, 0, 0, 1, 1, 1])
w_tmp = np.array([2.,3.])
b_tmp = 1.
dj_db_tmp, dj_dw_tmp = compute_gradient_logistic(X_tmp, y_tmp, w_tmp, b_tmp)
print(f"dj_db: {dj_db_tmp}" )
print(f"dj_dw: {dj_dw_tmp.tolist()}" )

3.2 梯度下降
实现上述公式(1),代码为:
def gradient_descent(X, y, w_in, b_in, alpha, num_iters):
"""
Performs batch gradient descent
Args:
X (ndarray (m,n) : Data, m examples with n features
y (ndarray (m,)) : target values
w_in (ndarray (n,)): Initial values of model parameters
b_in (scalar) : Initial values of model parameter
alpha (float) : Learning rate
num_iters (scalar) : number of iterations to run gradient descent
Returns:
w (ndarray (n,)) : Updated values of parameters
b (scalar) : Updated value of parameter
"""
# An array to store cost J and w's at each iteration primarily for graphing later
J_history = []
w = copy.deepcopy(w_in) #avoid modifying global w within function
b = b_in
for i in range(num_iters):
# Calculate the gradient and update the parameters
dj_db, dj_dw = compute_gradient_logistic(X, y, w, b)
# Update Parameters using w, b, alpha and gradient
w = w - alpha * dj_dw
b = b - alpha * dj_db
# Save cost J at each iteration
if i<100000: # prevent resource exhaustion
J_history.append( compute_cost_logistic(X, y, w, b) )
# Print cost every at intervals 10 times or as many iterations if < 10
if i% math.ceil(num_iters / 10) == 0:
print(f"Iteration {i:4d}: Cost {J_history[-1]} ")
return w, b, J_history #return final w,b and J history for graphing
运行一下:
w_tmp = np.zeros_like(X_train[0])
b_tmp = 0.
alph = 0.1
iters = 10000
w_out, b_out, _ = gradient_descent(X_train, y_train, w_tmp, b_tmp, alph, iters)
print(f"\nupdated parameters: w:{w_out}, b:{b_out}")

梯度下降的结果可视化:
fig,ax = plt.subplots(1,1,figsize=(5,4))
# plot the probability
plt_prob(ax, w_out, b_out)
# Plot the original data
ax.set_ylabel(r'$x_1$')
ax.set_xlabel(r'$x_0$')
ax.axis([0, 4, 0, 3.5])
plot_data(X_train,y_train,ax)
# Plot the decision boundary
x0 = -b_out/w_out[1]
x1 = -b_out/w_out[0]
ax.plot([0,x0],[x1,0], c=dlc["dlblue"], lw=1)
plt.show()
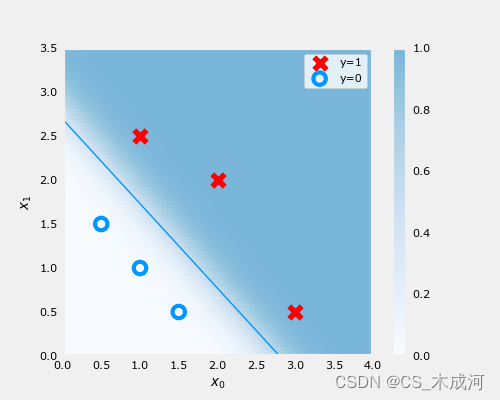
在上图中,阴影部分表示概率 y=1,决策边界是概率为0.5的直线。
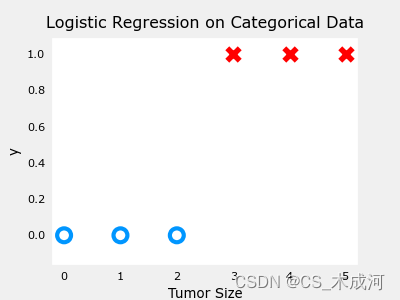
4. 数据集(单变量)
导入数据绘图可视化,此时参数为 w w w, b b b:
x_train = np.array([0., 1, 2, 3, 4, 5])
y_train = np.array([0, 0, 0, 1, 1, 1])
fig,ax = plt.subplots(1,1,figsize=(4,3))
plt_tumor_data(x_train, y_train, ax)
plt.show()
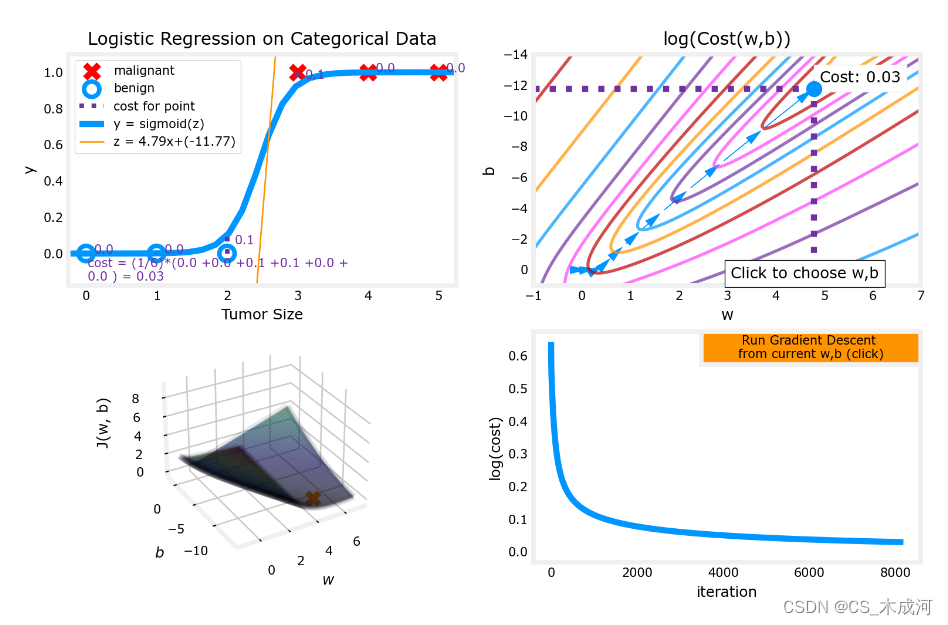
w_range = np.array([-1, 7])
b_range = np.array([1, -14])
quad = plt_quad_logistic( x_train, y_train, w_range, b_range )
附录
lab_utils_common.py 源码:
"""
lab_utils_common
contains common routines and variable definitions
used by all the labs in this week.
by contrast, specific, large plotting routines will be in separate files
and are generally imported into the week where they are used.
those files will import this file
"""
import copy
import math
import numpy as np
import matplotlib.pyplot as plt
from matplotlib.patches import FancyArrowPatch
from ipywidgets import Output
np.set_printoptions(precision=2)
dlc = dict(dlblue = '#0096ff', dlorange = '#FF9300', dldarkred='#C00000', dlmagenta='#FF40FF', dlpurple='#7030A0')
dlblue = '#0096ff'; dlorange = '#FF9300'; dldarkred='#C00000'; dlmagenta='#FF40FF'; dlpurple='#7030A0'
dlcolors = [dlblue, dlorange, dldarkred, dlmagenta, dlpurple]
plt.style.use('./deeplearning.mplstyle')
def sigmoid(z):
"""
Compute the sigmoid of z
Parameters
----------
z : array_like
A scalar or numpy array of any size.
Returns
-------
g : array_like
sigmoid(z)
"""
z = np.clip( z, -500, 500 ) # protect against overflow
g = 1.0/(1.0+np.exp(-z))
return g
##########################################################
# Regression Routines
##########################################################
def predict_logistic(X, w, b):
""" performs prediction """
return sigmoid(X @ w + b)
def predict_linear(X, w, b):
""" performs prediction """
return X @ w + b
def compute_cost_logistic(X, y, w, b, lambda_=0, safe=False):
"""
Computes cost using logistic loss, non-matrix version
Args:
X (ndarray): Shape (m,n) matrix of examples with n features
y (ndarray): Shape (m,) target values
w (ndarray): Shape (n,) parameters for prediction
b (scalar): parameter for prediction
lambda_ : (scalar, float) Controls amount of regularization, 0 = no regularization
safe : (boolean) True-selects under/overflow safe algorithm
Returns:
cost (scalar): cost
"""
m,n = X.shape
cost = 0.0
for i in range(m):
z_i = np.dot(X[i],w) + b #(n,)(n,) or (n,) ()
if safe: #avoids overflows
cost += -(y[i] * z_i ) + log_1pexp(z_i)
else:
f_wb_i = sigmoid(z_i) #(n,)
cost += -y[i] * np.log(f_wb_i) - (1 - y[i]) * np.log(1 - f_wb_i) # scalar
cost = cost/m
reg_cost = 0
if lambda_ != 0:
for j in range(n):
reg_cost += (w[j]**2) # scalar
reg_cost = (lambda_/(2*m))*reg_cost
return cost + reg_cost
def log_1pexp(x, maximum=20):
''' approximate log(1+exp^x)
https://stats.stackexchange.com/questions/475589/numerical-computation-of-cross-entropy-in-practice
Args:
x : (ndarray Shape (n,1) or (n,) input
out : (ndarray Shape matches x output ~= np.log(1+exp(x))
'''
out = np.zeros_like(x,dtype=float)
i = x <= maximum
ni = np.logical_not(i)
out[i] = np.log(1 + np.exp(x[i]))
out[ni] = x[ni]
return out
def compute_cost_matrix(X, y, w, b, logistic=False, lambda_=0, safe=True):
"""
Computes the cost using using matrices
Args:
X : (ndarray, Shape (m,n)) matrix of examples
y : (ndarray Shape (m,) or (m,1)) target value of each example
w : (ndarray Shape (n,) or (n,1)) Values of parameter(s) of the model
b : (scalar ) Values of parameter of the model
verbose : (Boolean) If true, print out intermediate value f_wb
Returns:
total_cost: (scalar) cost
"""
m = X.shape[0]
y = y.reshape(-1,1) # ensure 2D
w = w.reshape(-1,1) # ensure 2D
if logistic:
if safe: #safe from overflow
z = X @ w + b #(m,n)(n,1)=(m,1)
cost = -(y * z) + log_1pexp(z)
cost = np.sum(cost)/m # (scalar)
else:
f = sigmoid(X @ w + b) # (m,n)(n,1) = (m,1)
cost = (1/m)*(np.dot(-y.T, np.log(f)) - np.dot((1-y).T, np.log(1-f))) # (1,m)(m,1) = (1,1)
cost = cost[0,0] # scalar
else:
f = X @ w + b # (m,n)(n,1) = (m,1)
cost = (1/(2*m)) * np.sum((f - y)**2) # scalar
reg_cost = (lambda_/(2*m)) * np.sum(w**2) # scalar
total_cost = cost + reg_cost # scalar
return total_cost # scalar
def compute_gradient_matrix(X, y, w, b, logistic=False, lambda_=0):
"""
Computes the gradient using matrices
Args:
X : (ndarray, Shape (m,n)) matrix of examples
y : (ndarray Shape (m,) or (m,1)) target value of each example
w : (ndarray Shape (n,) or (n,1)) Values of parameters of the model
b : (scalar ) Values of parameter of the model
logistic: (boolean) linear if false, logistic if true
lambda_: (float) applies regularization if non-zero
Returns
dj_dw: (array_like Shape (n,1)) The gradient of the cost w.r.t. the parameters w
dj_db: (scalar) The gradient of the cost w.r.t. the parameter b
"""
m = X.shape[0]
y = y.reshape(-1,1) # ensure 2D
w = w.reshape(-1,1) # ensure 2D
f_wb = sigmoid( X @ w + b ) if logistic else X @ w + b # (m,n)(n,1) = (m,1)
err = f_wb - y # (m,1)
dj_dw = (1/m) * (X.T @ err) # (n,m)(m,1) = (n,1)
dj_db = (1/m) * np.sum(err) # scalar
dj_dw += (lambda_/m) * w # regularize # (n,1)
return dj_db, dj_dw # scalar, (n,1)
def gradient_descent(X, y, w_in, b_in, alpha, num_iters, logistic=False, lambda_=0, verbose=True):
"""
Performs batch gradient descent to learn theta. Updates theta by taking
num_iters gradient steps with learning rate alpha
Args:
X (ndarray): Shape (m,n) matrix of examples
y (ndarray): Shape (m,) or (m,1) target value of each example
w_in (ndarray): Shape (n,) or (n,1) Initial values of parameters of the model
b_in (scalar): Initial value of parameter of the model
logistic: (boolean) linear if false, logistic if true
lambda_: (float) applies regularization if non-zero
alpha (float): Learning rate
num_iters (int): number of iterations to run gradient descent
Returns:
w (ndarray): Shape (n,) or (n,1) Updated values of parameters; matches incoming shape
b (scalar): Updated value of parameter
"""
# An array to store cost J and w's at each iteration primarily for graphing later
J_history = []
w = copy.deepcopy(w_in) #avoid modifying global w within function
b = b_in
w = w.reshape(-1,1) #prep for matrix operations
y = y.reshape(-1,1)
for i in range(num_iters):
# Calculate the gradient and update the parameters
dj_db,dj_dw = compute_gradient_matrix(X, y, w, b, logistic, lambda_)
# Update Parameters using w, b, alpha and gradient
w = w - alpha * dj_dw
b = b - alpha * dj_db
# Save cost J at each iteration
if i<100000: # prevent resource exhaustion
J_history.append( compute_cost_matrix(X, y, w, b, logistic, lambda_) )
# Print cost every at intervals 10 times or as many iterations if < 10
if i% math.ceil(num_iters / 10) == 0:
if verbose: print(f"Iteration {i:4d}: Cost {J_history[-1]} ")
return w.reshape(w_in.shape), b, J_history #return final w,b and J history for graphing
def zscore_normalize_features(X):
"""
computes X, zcore normalized by column
Args:
X (ndarray): Shape (m,n) input data, m examples, n features
Returns:
X_norm (ndarray): Shape (m,n) input normalized by column
mu (ndarray): Shape (n,) mean of each feature
sigma (ndarray): Shape (n,) standard deviation of each feature
"""
# find the mean of each column/feature
mu = np.mean(X, axis=0) # mu will have shape (n,)
# find the standard deviation of each column/feature
sigma = np.std(X, axis=0) # sigma will have shape (n,)
# element-wise, subtract mu for that column from each example, divide by std for that column
X_norm = (X - mu) / sigma
return X_norm, mu, sigma
#check our work
#from sklearn.preprocessing import scale
#scale(X_orig, axis=0, with_mean=True, with_std=True, copy=True)
######################################################
# Common Plotting Routines
######################################################
def plot_data(X, y, ax, pos_label="y=1", neg_label="y=0", s=80, loc='best' ):
""" plots logistic data with two axis """
# Find Indices of Positive and Negative Examples
pos = y == 1
neg = y == 0
pos = pos.reshape(-1,) #work with 1D or 1D y vectors
neg = neg.reshape(-1,)
# Plot examples
ax.scatter(X[pos, 0], X[pos, 1], marker='x', s=s, c = 'red', label=pos_label)
ax.scatter(X[neg, 0], X[neg, 1], marker='o', s=s, label=neg_label, facecolors='none', edgecolors=dlblue, lw=3)
ax.legend(loc=loc)
ax.figure.canvas.toolbar_visible = False
ax.figure.canvas.header_visible = False
ax.figure.canvas.footer_visible = False
def plt_tumor_data(x, y, ax):
""" plots tumor data on one axis """
pos = y == 1
neg = y == 0
ax.scatter(x[pos], y[pos], marker='x', s=80, c = 'red', label="malignant")
ax.scatter(x[neg], y[neg], marker='o', s=100, label="benign", facecolors='none', edgecolors=dlblue,lw=3)
ax.set_ylim(-0.175,1.1)
ax.set_ylabel('y')
ax.set_xlabel('Tumor Size')
ax.set_title("Logistic Regression on Categorical Data")
ax.figure.canvas.toolbar_visible = False
ax.figure.canvas.header_visible = False
ax.figure.canvas.footer_visible = False
# Draws a threshold at 0.5
def draw_vthresh(ax,x):
""" draws a threshold """
ylim = ax.get_ylim()
xlim = ax.get_xlim()
ax.fill_between([xlim[0], x], [ylim[1], ylim[1]], alpha=0.2, color=dlblue)
ax.fill_between([x, xlim[1]], [ylim[1], ylim[1]], alpha=0.2, color=dldarkred)
ax.annotate("z >= 0", xy= [x,0.5], xycoords='data',
xytext=[30,5],textcoords='offset points')
d = FancyArrowPatch(
posA=(x, 0.5), posB=(x+3, 0.5), color=dldarkred,
arrowstyle='simple, head_width=5, head_length=10, tail_width=0.0',
)
ax.add_artist(d)
ax.annotate("z < 0", xy= [x,0.5], xycoords='data',
xytext=[-50,5],textcoords='offset points', ha='left')
f = FancyArrowPatch(
posA=(x, 0.5), posB=(x-3, 0.5), color=dlblue,
arrowstyle='simple, head_width=5, head_length=10, tail_width=0.0',
)
ax.add_artist(f)
plt_quad_logistic.py 源码:
"""
plt_quad_logistic.py
interactive plot and supporting routines showing logistic regression
"""
import time
from matplotlib import cm
import matplotlib.colors as colors
from matplotlib.gridspec import GridSpec
from matplotlib.widgets import Button
from matplotlib.patches import FancyArrowPatch
from ipywidgets import Output
from lab_utils_common import np, plt, dlc, dlcolors, sigmoid, compute_cost_matrix, gradient_descent
# for debug
#output = Output() # sends hidden error messages to display when using widgets
#display(output)
class plt_quad_logistic:
''' plots a quad plot showing logistic regression '''
# pylint: disable=too-many-instance-attributes
# pylint: disable=too-many-locals
# pylint: disable=missing-function-docstring
# pylint: disable=attribute-defined-outside-init
def __init__(self, x_train,y_train, w_range, b_range):
# setup figure
fig = plt.figure( figsize=(10,6))
fig.canvas.toolbar_visible = False
fig.canvas.header_visible = False
fig.canvas.footer_visible = False
fig.set_facecolor('#ffffff') #white
gs = GridSpec(2, 2, figure=fig)
ax0 = fig.add_subplot(gs[0, 0])
ax1 = fig.add_subplot(gs[0, 1])
ax2 = fig.add_subplot(gs[1, 0], projection='3d')
ax3 = fig.add_subplot(gs[1,1])
pos = ax3.get_position().get_points() ##[[lb_x,lb_y], [rt_x, rt_y]]
h = 0.05
width = 0.2
axcalc = plt.axes([pos[1,0]-width, pos[1,1]-h, width, h]) #lx,by,w,h
ax = np.array([ax0, ax1, ax2, ax3, axcalc])
self.fig = fig
self.ax = ax
self.x_train = x_train
self.y_train = y_train
self.w = 0. #initial point, non-array
self.b = 0.
# initialize subplots
self.dplot = data_plot(ax[0], x_train, y_train, self.w, self.b)
self.con_plot = contour_and_surface_plot(ax[1], ax[2], x_train, y_train, w_range, b_range, self.w, self.b)
self.cplot = cost_plot(ax[3])
# setup events
self.cid = fig.canvas.mpl_connect('button_press_event', self.click_contour)
self.bcalc = Button(axcalc, 'Run Gradient Descent \nfrom current w,b (click)', color=dlc["dlorange"])
self.bcalc.on_clicked(self.calc_logistic)
# @output.capture() # debug
def click_contour(self, event):
''' called when click in contour '''
if event.inaxes == self.ax[1]: #contour plot
self.w = event.xdata
self.b = event.ydata
self.cplot.re_init()
self.dplot.update(self.w, self.b)
self.con_plot.update_contour_wb_lines(self.w, self.b)
self.con_plot.path.re_init(self.w, self.b)
self.fig.canvas.draw()
# @output.capture() # debug
def calc_logistic(self, event):
''' called on run gradient event '''
for it in [1, 8,16,32,64,128,256,512,1024,2048,4096]:
w, self.b, J_hist = gradient_descent(self.x_train.reshape(-1,1), self.y_train.reshape(-1,1),
np.array(self.w).reshape(-1,1), self.b, 0.1, it,
logistic=True, lambda_=0, verbose=False)
self.w = w[0,0]
self.dplot.update(self.w, self.b)
self.con_plot.update_contour_wb_lines(self.w, self.b)
self.con_plot.path.add_path_item(self.w,self.b)
self.cplot.add_cost(J_hist)
time.sleep(0.3)
self.fig.canvas.draw()
class data_plot:
''' handles data plot '''
# pylint: disable=missing-function-docstring
# pylint: disable=attribute-defined-outside-init
def __init__(self, ax, x_train, y_train, w, b):
self.ax = ax
self.x_train = x_train
self.y_train = y_train
self.m = x_train.shape[0]
self.w = w
self.b = b
self.plt_tumor_data()
self.draw_logistic_lines(firsttime=True)
self.mk_cost_lines(firsttime=True)
self.ax.autoscale(enable=False) # leave plot scales the same after initial setup
def plt_tumor_data(self):
x = self.x_train
y = self.y_train
pos = y == 1
neg = y == 0
self.ax.scatter(x[pos], y[pos], marker='x', s=80, c = 'red', label="malignant")
self.ax.scatter(x[neg], y[neg], marker='o', s=100, label="benign", facecolors='none',
edgecolors=dlc["dlblue"],lw=3)
self.ax.set_ylim(-0.175,1.1)
self.ax.set_ylabel('y')
self.ax.set_xlabel('Tumor Size')
self.ax.set_title("Logistic Regression on Categorical Data")
def update(self, w, b):
self.w = w
self.b = b
self.draw_logistic_lines()
self.mk_cost_lines()
def draw_logistic_lines(self, firsttime=False):
if not firsttime:
self.aline[0].remove()
self.bline[0].remove()
self.alegend.remove()
xlim = self.ax.get_xlim()
x_hat = np.linspace(*xlim, 30)
y_hat = sigmoid(np.dot(x_hat.reshape(-1,1), self.w) + self.b)
self.aline = self.ax.plot(x_hat, y_hat, color=dlc["dlblue"],
label="y = sigmoid(z)")
f_wb = np.dot(x_hat.reshape(-1,1), self.w) + self.b
self.bline = self.ax.plot(x_hat, f_wb, color=dlc["dlorange"], lw=1,
label=f"z = {np.squeeze(self.w):0.2f}x+({self.b:0.2f})")
self.alegend = self.ax.legend(loc='upper left')
def mk_cost_lines(self, firsttime=False):
''' makes vertical cost lines'''
if not firsttime:
for artist in self.cost_items:
artist.remove()
self.cost_items = []
cstr = f"cost = (1/{self.m})*("
ctot = 0
label = 'cost for point'
addedbreak = False
for p in zip(self.x_train,self.y_train):
f_wb_p = sigmoid(self.w*p[0]+self.b)
c_p = compute_cost_matrix(p[0].reshape(-1,1), p[1],np.array(self.w), self.b, logistic=True, lambda_=0, safe=True)
c_p_txt = c_p
a = self.ax.vlines(p[0], p[1],f_wb_p, lw=3, color=dlc["dlpurple"], ls='dotted', label=label)
label='' #just one
cxy = [p[0], p[1] + (f_wb_p-p[1])/2]
b = self.ax.annotate(f'{c_p_txt:0.1f}', xy=cxy, xycoords='data',color=dlc["dlpurple"],
xytext=(5, 0), textcoords='offset points')
cstr += f"{c_p_txt:0.1f} +"
if len(cstr) > 38 and addedbreak is False:
cstr += "\n"
addedbreak = True
ctot += c_p
self.cost_items.extend((a,b))
ctot = ctot/(len(self.x_train))
cstr = cstr[:-1] + f") = {ctot:0.2f}"
## todo.. figure out how to get this textbox to extend to the width of the subplot
c = self.ax.text(0.05,0.02,cstr, transform=self.ax.transAxes, color=dlc["dlpurple"])
self.cost_items.append(c)
class contour_and_surface_plot:
''' plots combined in class as they have similar operations '''
# pylint: disable=missing-function-docstring
# pylint: disable=attribute-defined-outside-init
def __init__(self, axc, axs, x_train, y_train, w_range, b_range, w, b):
self.x_train = x_train
self.y_train = y_train
self.axc = axc
self.axs = axs
#setup useful ranges and common linspaces
b_space = np.linspace(*b_range, 100)
w_space = np.linspace(*w_range, 100)
# get cost for w,b ranges for contour and 3D
tmp_b,tmp_w = np.meshgrid(b_space,w_space)
z = np.zeros_like(tmp_b)
for i in range(tmp_w.shape[0]):
for j in range(tmp_w.shape[1]):
z[i,j] = compute_cost_matrix(x_train.reshape(-1,1), y_train, tmp_w[i,j], tmp_b[i,j],
logistic=True, lambda_=0, safe=True)
if z[i,j] == 0:
z[i,j] = 1e-9
### plot contour ###
CS = axc.contour(tmp_w, tmp_b, np.log(z),levels=12, linewidths=2, alpha=0.7,colors=dlcolors)
axc.set_title('log(Cost(w,b))')
axc.set_xlabel('w', fontsize=10)
axc.set_ylabel('b', fontsize=10)
axc.set_xlim(w_range)
axc.set_ylim(b_range)
self.update_contour_wb_lines(w, b, firsttime=True)
axc.text(0.7,0.05,"Click to choose w,b", bbox=dict(facecolor='white', ec = 'black'), fontsize = 10,
transform=axc.transAxes, verticalalignment = 'center', horizontalalignment= 'center')
#Surface plot of the cost function J(w,b)
axs.plot_surface(tmp_w, tmp_b, z, cmap = cm.jet, alpha=0.3, antialiased=True)
axs.plot_wireframe(tmp_w, tmp_b, z, color='k', alpha=0.1)
axs.set_xlabel("$w$")
axs.set_ylabel("$b$")
axs.zaxis.set_rotate_label(False)
axs.xaxis.set_pane_color((1.0, 1.0, 1.0, 0.0))
axs.yaxis.set_pane_color((1.0, 1.0, 1.0, 0.0))
axs.zaxis.set_pane_color((1.0, 1.0, 1.0, 0.0))
axs.set_zlabel("J(w, b)", rotation=90)
axs.view_init(30, -120)
axs.autoscale(enable=False)
axc.autoscale(enable=False)
self.path = path(self.w,self.b, self.axc) # initialize an empty path, avoids existance check
def update_contour_wb_lines(self, w, b, firsttime=False):
self.w = w
self.b = b
cst = compute_cost_matrix(self.x_train.reshape(-1,1), self.y_train, np.array(self.w), self.b,
logistic=True, lambda_=0, safe=True)
# remove lines and re-add on contour plot and 3d plot
if not firsttime:
for artist in self.dyn_items:
artist.remove()
a = self.axc.scatter(self.w, self.b, s=100, color=dlc["dlblue"], zorder= 10, label="cost with \ncurrent w,b")
b = self.axc.hlines(self.b, self.axc.get_xlim()[0], self.w, lw=4, color=dlc["dlpurple"], ls='dotted')
c = self.axc.vlines(self.w, self.axc.get_ylim()[0] ,self.b, lw=4, color=dlc["dlpurple"], ls='dotted')
d = self.axc.annotate(f"Cost: {cst:0.2f}", xy= (self.w, self.b), xytext = (4,4), textcoords = 'offset points',
bbox=dict(facecolor='white'), size = 10)
#Add point in 3D surface plot
e = self.axs.scatter3D(self.w, self.b, cst , marker='X', s=100)
self.dyn_items = [a,b,c,d,e]
class cost_plot:
""" manages cost plot for plt_quad_logistic """
# pylint: disable=missing-function-docstring
# pylint: disable=attribute-defined-outside-init
def __init__(self,ax):
self.ax = ax
self.ax.set_ylabel("log(cost)")
self.ax.set_xlabel("iteration")
self.costs = []
self.cline = self.ax.plot(0,0, color=dlc["dlblue"])
def re_init(self):
self.ax.clear()
self.__init__(self.ax)
def add_cost(self,J_hist):
self.costs.extend(J_hist)
self.cline[0].remove()
self.cline = self.ax.plot(self.costs)
class path:
''' tracks paths during gradient descent on contour plot '''
# pylint: disable=missing-function-docstring
# pylint: disable=attribute-defined-outside-init
def __init__(self, w, b, ax):
''' w, b at start of path '''
self.path_items = []
self.w = w
self.b = b
self.ax = ax
def re_init(self, w, b):
for artist in self.path_items:
artist.remove()
self.path_items = []
self.w = w
self.b = b
def add_path_item(self, w, b):
a = FancyArrowPatch(
posA=(self.w, self.b), posB=(w, b), color=dlc["dlblue"],
arrowstyle='simple, head_width=5, head_length=10, tail_width=0.0',
)
self.ax.add_artist(a)
self.path_items.append(a)
self.w = w
self.b = b
#-----------
# related to the logistic gradient descent lab
#----------
def truncate_colormap(cmap, minval=0.0, maxval=1.0, n=100):
""" truncates color map """
new_cmap = colors.LinearSegmentedColormap.from_list(
'trunc({n},{a:.2f},{b:.2f})'.format(n=cmap.name, a=minval, b=maxval),
cmap(np.linspace(minval, maxval, n)))
return new_cmap
def plt_prob(ax, w_out,b_out):
""" plots a decision boundary but include shading to indicate the probability """
#setup useful ranges and common linspaces
x0_space = np.linspace(0, 4 , 100)
x1_space = np.linspace(0, 4 , 100)
# get probability for x0,x1 ranges
tmp_x0,tmp_x1 = np.meshgrid(x0_space,x1_space)
z = np.zeros_like(tmp_x0)
for i in range(tmp_x0.shape[0]):
for j in range(tmp_x1.shape[1]):
z[i,j] = sigmoid(np.dot(w_out, np.array([tmp_x0[i,j],tmp_x1[i,j]])) + b_out)
cmap = plt.get_cmap('Blues')
new_cmap = truncate_colormap(cmap, 0.0, 0.5)
pcm = ax.pcolormesh(tmp_x0, tmp_x1, z,
norm=cm.colors.Normalize(vmin=0, vmax=1),
cmap=new_cmap, shading='nearest', alpha = 0.9)
ax.figure.colorbar(pcm, ax=ax)
























 3963
3963











 被折叠的 条评论
为什么被折叠?
被折叠的 条评论
为什么被折叠?










