去年上课的时候老师简要提到过,但是当时笔记太简略了orz
https://scikit-learn.org/stable/auto_examples/classification/plot_lda_qda.html
这里记录一下sklearn给的示例代码,以及一些相关的笔记
import matplotlib.pyplot as plt
import matplotlib as mpl
from matplotlib import colors
'''
------------------- colormap -------------------
'''
# 设置颜色,'red_blue_classes' 只是标签而已
# 在后续画图的时候可以指定使用这一标签对应的染色方法
# segmentdata argument is a dictionary with a red, green and blue entries.
# Each entry should be a list of x, y0, y1 tuples, forming rows in a table.
# Entries for alpha are optional. y0 y1应该是颜色变化的范围,x在行之间的变化标识位置
cmap = colors.LinearSegmentedColormap(
"red_blue_classes",
{
"red": [(0, 1, 1), (1, 0.7, 0.7)],
"green": [(0, 0.7, 0.7), (1, 0.7, 0.7)],
"blue": [(0, 0.7, 0.7), (1, 1, 1)],
}
)
# Register a new colormap to be accessed by name
# 之后可以按标签调取调色,与colors.LinearSegmentedColormap搭配使用
plt.cm.register_cmap(cmap=cmap)
import numpy as np
'''
------------------- Datasets generation function -------------------
'''
def dataset_fixed_cov():
"""Generate 2 Gaussians samples with the same covariance matrix"""
n, dim = 300, 2
np.random.seed(0)
C = np.array([[0.0, -0.23], [0.83, 0.23]])
X = np.r_[
np.dot(np.random.randn(n, dim), C),
np.dot(np.random.randn(n, dim), C) + np.array([1, 1]),
]
y = np.hstack((np.zeros(n), np.ones(n)))
return X, y
def dataset_cov():
"""Generate 2 Gaussians samples with different covariance matrices"""
n, dim = 300, 2
np.random.seed(0)
C = np.array([[0.0, -1.0], [2.5, 0.7]]) * 2.0
# 简单通俗地来说,就是将一些slice object进行沿第一轴连接(通俗一点的说法是row-wise)连接。
# 但是,row-wise这个说法其实是有点含糊,因为这个预设了矩阵这种形象。因为行和列仅在2维的情况下才有意义。
# 在更广义的多维数组的话语中,已经没有行和列这种概念。所以说‘along the first axis’是严谨而通用的。
X = np.r_[
# dot()返回的是两个数组的点积(dot product)
# 如果是二维数组(矩阵)之间的运算,则得到的是矩阵积(mastrix product)。
# https://www.cnblogs.com/luhuan/p/7925790.html
np.dot(np.random.randn(n, dim), C),
np.dot(np.random.randn(n, dim), C.T) + np.array([1, 4]),
]
# 按水平方向(列顺序)堆叠数组构成一个新的数组
y = np.hstack((np.zeros(n), np.ones(n)))
return X, y
'''
------------------- Plot Functions -------------------
'''
from scipy import linalg
def plot_data(lda, X, y, y_pred, fig_index):
# 按subfigure的序号给x y位置索引
splot = plt.subplot(2, 2, fig_index)
if fig_index == 1:
plt.title("Linear Discriminant Analysis")
plt.ylabel("Data with\n fixed covariance")
elif fig_index == 2:
plt.title("Quadratic Discriminant Analysis")
elif fig_index == 3:
plt.ylabel("Data with\n varying covariances")
tp = y == y_pred # True Positive
# True Positive(简称TP):判断为正,且实际为正。
# 但是注意这里,只要predict是对的就行,(TP/TN)
'''
yy = np.array([0, 1, 0, 1])
yyy = np.array([0, 0, 1, 1])
a = yy==yyy
print('a', a)
print('yy==0', a[yy==0])
print('yy==1', a[yy==1])
对应 yy == 0的位置,查看其在bool列表中对应的是T/F
a [ True False False True]
yy==0 [ True False]
yy==1 [False True]
'''
# 因此tp0是原类别是0的点对其类别的判断是否正确,tp1是原类别是1的点对其类别的判断是否正确
tp0, tp1 = tp[y == 0], tp[y == 1]
# X0是类别0对应的feature, X1是类别1对应的feature
X0, X1 = X[y == 0], X[y == 1]
#
X0_tp, X0_fp = X0[tp0], X0[~tp0]
X1_tp, X1_fp = X1[tp1], X1[~tp1]
# class 0: dots
# 类别0,判断对的·,判断错的×
plt.scatter(X0_tp[:, 0], X0_tp[:, 1], marker=".", color="red")
plt.scatter(X0_fp[:, 0], X0_fp[:, 1], marker="x", s=20, color="#990000") # dark red
# class 1: dots
# 类别0,判断对的·,判断错的×
plt.scatter(X1_tp[:, 0], X1_tp[:, 1], marker=".", color="blue")
plt.scatter(X1_fp[:, 0], X1_fp[:, 1], marker="x", s=20, color="#000099") # dark blue
# class 0 and 1 : areas
nx, ny = 200, 100
x_min, x_max = plt.xlim()
y_min, y_max = plt.ylim()
# X, Y = np.meshgrid(x, y)
# 代表的是将x中每一个数据和y中每一个数据组合生成很多点, 然后将这些点的x坐标放入到X中, y坐标放入Y中, 并且相应位置是对应的
# np.linspace(start = 0, stop = 100, num = 5) 通过定义均匀间隔创建数值序列
xx, yy = np.meshgrid(np.linspace(x_min, x_max, nx), np.linspace(y_min, y_max, ny))
# xx: (100, 200)
# ravel让多维数组变成一维数组
# np.c_是按行连接两个矩阵, 就是把两矩阵左右相加, 要求行数相等
# predict_proba(X) Estimate probability.
# Z: (20000, 2)
Z = lda.predict_proba(np.c_[xx.ravel(), yy.ravel()])
Z = Z[:, 1].reshape(xx.shape)
# Z: (100, 200)
# plt.pcolormesh(x1, x2, y_predict.reshape(x1.shape), cmap=cm_light)
# plt.pcolormesh()会根据y_predict的结果自动在cmap里选择颜色
plt.pcolormesh(xx, yy, Z, cmap="red_blue_classes", norm=colors.Normalize(0.0, 1.0), zorder=0)
# plt.contour是python中用于画等高线的函数
plt.contour(xx, yy, Z, [0.5], linewidths=2.0, colors="white")
# means
# means_: array-like of shape (n_classes, n_features)
# Class-wise means.
'''
在iris数据集中,类似的可以得到
# [[-1.01457897 0.85326268 -1.30498732 -1.25489349]
# [ 0.11228223 -0.66143204 0.28532388 0.1667341 ]
# [ 0.90229674 -0.19183064 1.01966344 1.08815939]]
显然,每一行是一个类别,每一列是该feature对应的中心点
'''
# 在这里,因为只有两个类别,且只有两个feature,所以得到的是一个2×2的矩阵
# 这里是第一个类别的中心点,画星星
plt.plot(
lda.means_[0][0],
lda.means_[0][1],
"*",
color="yellow",
markersize=15,
markeredgecolor="grey",
)
# 这里是第二个类别的中心点,也画星星
plt.plot(
lda.means_[1][0],
lda.means_[1][1],
"*",
color="yellow",
markersize=15,
markeredgecolor="grey",
)
return splot
def plot_ellipse(splot, mean, cov, color):
# 这里是计算的部分,参考LDA推导
# cov 2×2矩阵
# 求解复 Hermitian 或实对称矩阵的标准或广义特征值问题
# Solve a standard or generalized eigenvalue problem for a complex Hermitian or real symmetric matrix.
v, w = linalg.eigh(cov)
u = w[0] / linalg.norm(w[0])
angle = np.arctan(u[1] / u[0])
angle = 180 * angle / np.pi # convert to degrees
# filled Gaussian at 2 standard deviation
ell = mpl.patches.Ellipse(
mean,
2 * v[0] ** 0.5,
2 * v[1] ** 0.5,
angle=180 + angle,
facecolor=color,
edgecolor="black",
linewidth=2,
)
ell.set_clip_box(splot.bbox)
ell.set_alpha(0.2)
splot.add_artist(ell)
splot.set_xticks(())
splot.set_yticks(())
def plot_lda_cov(lda, splot):
plot_ellipse(splot, lda.means_[0], lda.covariance_, "red")
plot_ellipse(splot, lda.means_[1], lda.covariance_, "blue")
def plot_qda_cov(qda, splot):
plot_ellipse(splot, qda.means_[0], qda.covariance_[0], "red")
plot_ellipse(splot, qda.means_[1], qda.covariance_[1], "blue")
'''
------------------- Plot -------------------
'''
plt.figure(figsize=(8, 6), facecolor="white")
plt.suptitle(
"Linear Discriminant Analysis vs Quadratic Discriminant Analysis",
y=0.98,
fontsize=10,
)
from sklearn.discriminant_analysis import LinearDiscriminantAnalysis
from sklearn.discriminant_analysis import QuadraticDiscriminantAnalysis
for i, (X, y) in enumerate([dataset_fixed_cov(), dataset_cov()]):
# Linear Discriminant Analysis
lda = LinearDiscriminantAnalysis(solver="svd", store_covariance=True)
y_pred = lda.fit(X, y).predict(X)
splot = plot_data(lda, X, y, y_pred, fig_index=2 * i + 1)
plot_lda_cov(lda, splot)
plt.axis("tight")
# Quadratic Discriminant Analysis
qda = QuadraticDiscriminantAnalysis(store_covariance=True)
y_pred = qda.fit(X, y).predict(X)
splot = plot_data(qda, X, y, y_pred, fig_index=2 * i + 2)
plot_qda_cov(qda, splot)
plt.axis("tight")
plt.tight_layout()
plt.subplots_adjust(top=0.92)
plt.show()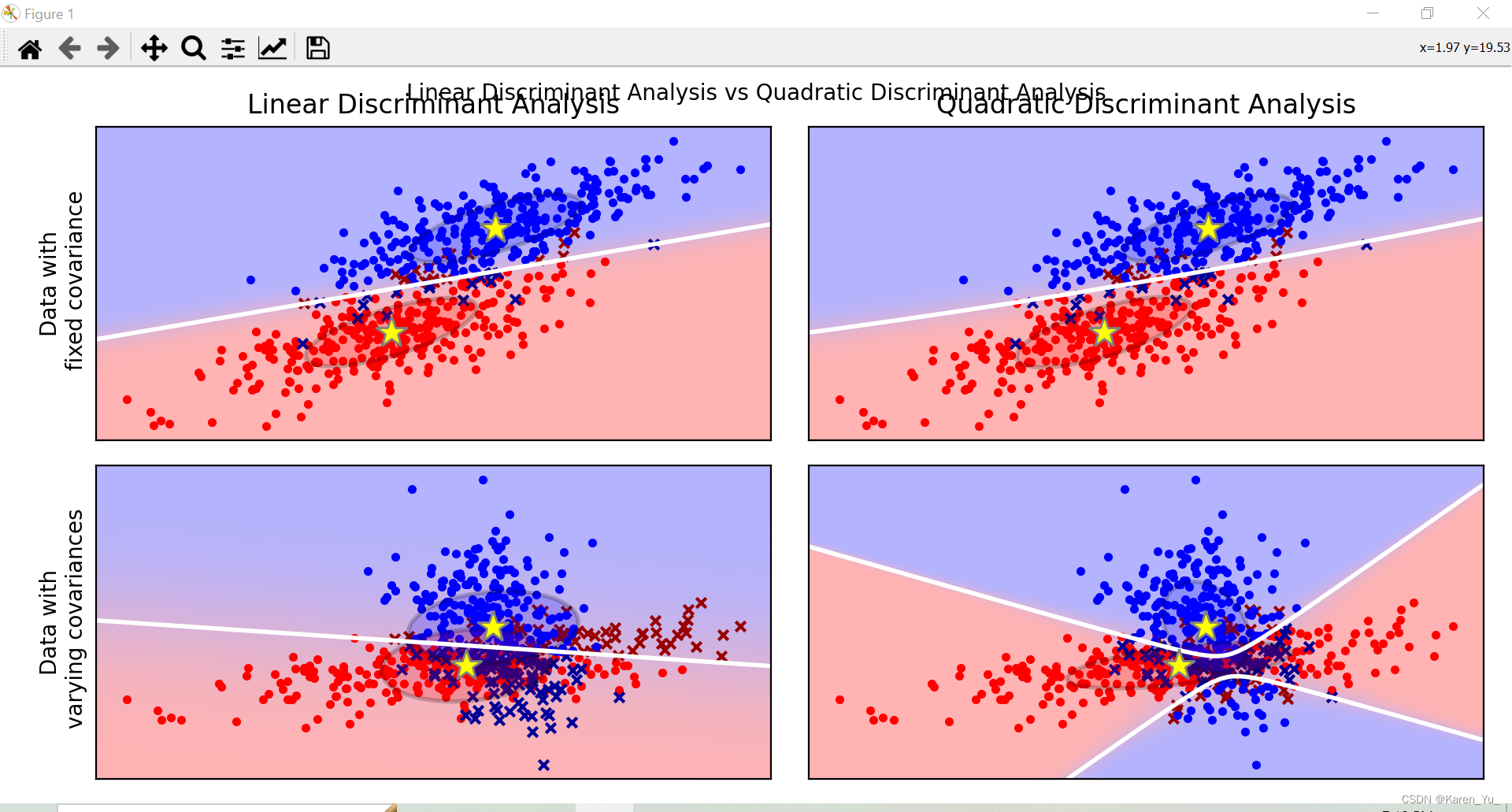
其他示例代码:
https://machinelearningmastery.com/linear-discriminant-analysis-with-python/
一些今天跟chatgpt的聊天:
🐰: What is Linear Discriminant Analysis? Is it a classification method?
💮:
Yes, Linear Discriminant Analysis (LDA) is a classification method commonly used in machine learning and statistics. It's a supervised learning algorithm that is used to find a linear combination of features that can distinguish between two or more classes in a dataset.
LDA is used for dimensionality reduction and classification tasks. It's particularly useful when dealing with high-dimensional data where the number of features is larger than the number of samples. The goal of LDA is to project the data onto a lower-dimensional space while still maintaining as much of the discriminatory information as possible.
LDA works by maximizing the between-class variance while minimizing the within-class variance. It finds a linear transformation that maximizes the ratio of between-class variance to within-class variance. This transformation is then used to project the data onto a lower-dimensional space, where classification can be performed using simple linear models such as logistic regression.
Overall, LDA is a powerful and widely used classification method that can help improve the accuracy of classification tasks, particularly when dealing with high-dimensional data.
🐰: Yes, I also read that LDA has been used for dimensionality reduction, this make me a bit confused, I once thought it's similar to PCA.
💮:
You're correct, LDA can also be used for dimensionality reduction. While LDA and PCA (Principal Component Analysis) are both techniques used for dimensionality reduction, they have different objectives and work differently.
PCA is an unsupervised learning technique that seeks to find the directions of maximum variance in the data. The idea is to project the data onto a lower-dimensional space by selecting the principal components that capture the most variation in the data. The principal components are not necessarily related to the class labels or any other external information, they only capture the overall structure of the data.
In contrast, LDA is a supervised learning technique that aims to find a lower-dimensional representation of the data that maximizes the separation between classes. Unlike PCA, LDA takes into account the class labels of the data and tries to find the projection that best separates the classes. Therefore, LDA can be seen as a technique that combines dimensionality reduction with classification.
In summary, while both techniques can be used for dimensionality reduction, they have different objectives and should be used in different situations. PCA is best used for data compression and visualization, while LDA is best used when the class labels of the data are known and classification is the ultimate goal.
🐰: I also find there is something called Fisher Discriminant Analysis, but the defination is quite similar to Linear Discriminant. So, what's the relationship between them?
💮:
Fisher Discriminant Analysis (FDA) is another name for Linear Discriminant Analysis (LDA). In fact, Fisher originally introduced LDA as a method for linear discriminant analysis, which is why you may see the terms used interchangeably in the literature.
The reason why LDA is also known as Fisher Discriminant Analysis is that it was developed by Ronald Fisher, a famous statistician and geneticist. Fisher introduced LDA in his seminal paper "The Use of Multiple Measurements in Taxonomic Problems" in 1936, where he presented a method for finding a linear combination of variables that best separates two or more classes.
So, in short, Fisher Discriminant Analysis and Linear Discriminant Analysis are the same thing, with LDA being the more commonly used term today.
小结:
LDA和FDA是一个东西,因为是Fisher这个人搞出来的,因此会看到两个名字混用
LDA主要用来降维和分类,其逻辑是以已有的类别标签为依据,寻找特征的线性组合,这一线性组合刚好可以区分两个或多个类,从而达到将数据投射到低维空间,并保留尽可能多的信息的目的,本质上就是线性变换
LDA和PCA都可以用来降维,但是两者本质不同
- 方法不同,LDA是supervised learning,类别标签是指定的,PCA是unsupervised learning, 不在乎类别标签,只关注overall structure of the data(类似于LDA会关注到有些地方是飞地,PCA只会觉得离得近的才是你的)
- 适用范围:LDA多用于类别标签已知且最终目标是分类,PCA多用于数据压缩和可视化(一个本身就是分类,一个只是进行下一步操作的手段)
类似的方法用在iris上再来一遍
from sklearn.datasets import load_iris
from matplotlib import colors
import matplotlib.pyplot as plt
import matplotlib as mpl
from scipy import linalg
import numpy as np
iris = load_iris()
X = iris.data[:, 2:4]
y = iris.target
cmap = colors.LinearSegmentedColormap(
"color_map",
{
"red": [(0, 1, 1), (1, 0.7, 0.7)],
"green": [(0, 0.7, 0.7), (1, 0.7, 0.7)],
"blue": [(0, 0.7, 0.7), (1, 1, 1)],
}
)
plt.cm.register_cmap(cmap=cmap)
def plot_data(lda, X, y, y_pred, fig_index):
# 按subfigure的序号给x y位置索引
splot = plt.subplot(1, 2, fig_index)
if fig_index == 1:
plt.title("Linear Discriminant Analysis")
plt.ylabel("Iris")
elif fig_index == 2:
plt.title("Quadratic Discriminant Analysis")
tp = y == y_pred # True Positive
# 因此tp0是原类别是0的点对其类别的判断是否正确,tp1是原类别是1的点对其类别的判断是否正确
tp0, tp1 = tp[y == 0], tp[y == 1]
# X0是类别0对应的feature, X1是类别1对应的feature
X0, X1 = X[y == 0], X[y == 1]
#
X0_tp, X0_fp = X0[tp0], X0[~tp0]
X1_tp, X1_fp = X1[tp1], X1[~tp1]
# class 0: dots
# 类别0,判断对的·,判断错的×
plt.scatter(X0_tp[:, 0], X0_tp[:, 1], marker=".", color="red")
plt.scatter(X0_fp[:, 0], X0_fp[:, 1], marker="x", s=20, color="#990000") # dark red
# class 1: dots
# 类别0,判断对的·,判断错的×
plt.scatter(X1_tp[:, 0], X1_tp[:, 1], marker=".", color="blue")
plt.scatter(X1_fp[:, 0], X1_fp[:, 1], marker="x", s=20, color="#000099") # dark blue
# class 0 and 1 : areas
nx, ny = 200, 100
x_min, x_max = plt.xlim()
y_min, y_max = plt.ylim()
xx, yy = np.meshgrid(np.linspace(x_min, x_max, nx), np.linspace(y_min, y_max, ny))
# xx: (100, 200)
# ravel让多维数组变成一维数组
# np.c_是按行连接两个矩阵, 就是把两矩阵左右相加, 要求行数相等
# predict_proba(X) Estimate probability.
# Z: (20000, 2)
Z = lda.predict_proba(np.c_[xx.ravel(), yy.ravel()])
Z = Z[:, 1].reshape(xx.shape)
# Z: (100, 200)
# plt.pcolormesh(x1, x2, y_predict.reshape(x1.shape), cmap=cm_light)
# plt.pcolormesh()会根据y_predict的结果自动在cmap里选择颜色
plt.pcolormesh(xx, yy, Z, cmap="color_map", norm=colors.Normalize(0.0, 1.0), zorder=0)
# plt.contour是python中用于画等高线的函数
plt.contour(xx, yy, Z, [0.5], linewidths=2.0, colors="white")
# means
# means_: array-like of shape (n_classes, n_features)
# Class-wise means.
# 这里是第一个类别的中心点,画星星
plt.plot(
lda.means_[0][0],
lda.means_[0][1],
"*",
color="yellow",
markersize=15,
markeredgecolor="grey",
)
# 这里是第二个类别的中心点,也画星星
plt.plot(
lda.means_[1][0],
lda.means_[1][1],
"*",
color="yellow",
markersize=15,
markeredgecolor="grey",
)
return splot
def plot_ellipse(splot, mean, cov, color):
# 这里是计算的部分,参考LDA推导
# cov 2×2矩阵
# 求解复 Hermitian 或实对称矩阵的标准或广义特征值问题
# Solve a standard or generalized eigenvalue problem for a complex Hermitian or real symmetric matrix.
v, w = linalg.eigh(cov)
u = w[0] / linalg.norm(w[0])
angle = np.arctan(u[1] / u[0])
angle = 180 * angle / np.pi # convert to degrees
# filled Gaussian at 2 standard deviation
ell = mpl.patches.Ellipse(
mean,
2 * v[0] ** 0.5,
2 * v[1] ** 0.5,
angle=180 + angle,
facecolor=color,
edgecolor="black",
linewidth=2,
)
ell.set_clip_box(splot.bbox)
ell.set_alpha(0.2)
splot.add_artist(ell)
splot.set_xticks(())
splot.set_yticks(())
def plot_lda_cov(lda, splot):
plot_ellipse(splot, lda.means_[0], lda.covariance_, "red")
plot_ellipse(splot, lda.means_[1], lda.covariance_, "blue")
def plot_qda_cov(qda, splot):
plot_ellipse(splot, qda.means_[0], qda.covariance_[0], "red")
plot_ellipse(splot, qda.means_[1], qda.covariance_[1], "blue")
'''
------------------- Plot -------------------
'''
plt.figure(figsize=(8, 6), facecolor="white")
plt.suptitle(
"Linear Discriminant Analysis vs Quadratic Discriminant Analysis",
y=0.98,
fontsize=10,
)
from sklearn.discriminant_analysis import LinearDiscriminantAnalysis
from sklearn.discriminant_analysis import QuadraticDiscriminantAnalysis
# Linear Discriminant Analysis
lda = LinearDiscriminantAnalysis(solver="svd", store_covariance=True)
y_pred = lda.fit(X, y).predict(X)
splot = plot_data(lda, X, y, y_pred, fig_index=2 * 0 + 1)
plot_lda_cov(lda, splot)
plt.axis("tight")
# Quadratic Discriminant Analysis
qda = QuadraticDiscriminantAnalysis(store_covariance=True)
y_pred = qda.fit(X, y).predict(X)
splot = plot_data(qda, X, y, y_pred, fig_index=2 * 0 + 2)
plot_qda_cov(qda, splot)
plt.axis("tight")
plt.tight_layout()
plt.subplots_adjust(top=0.92)
plt.show()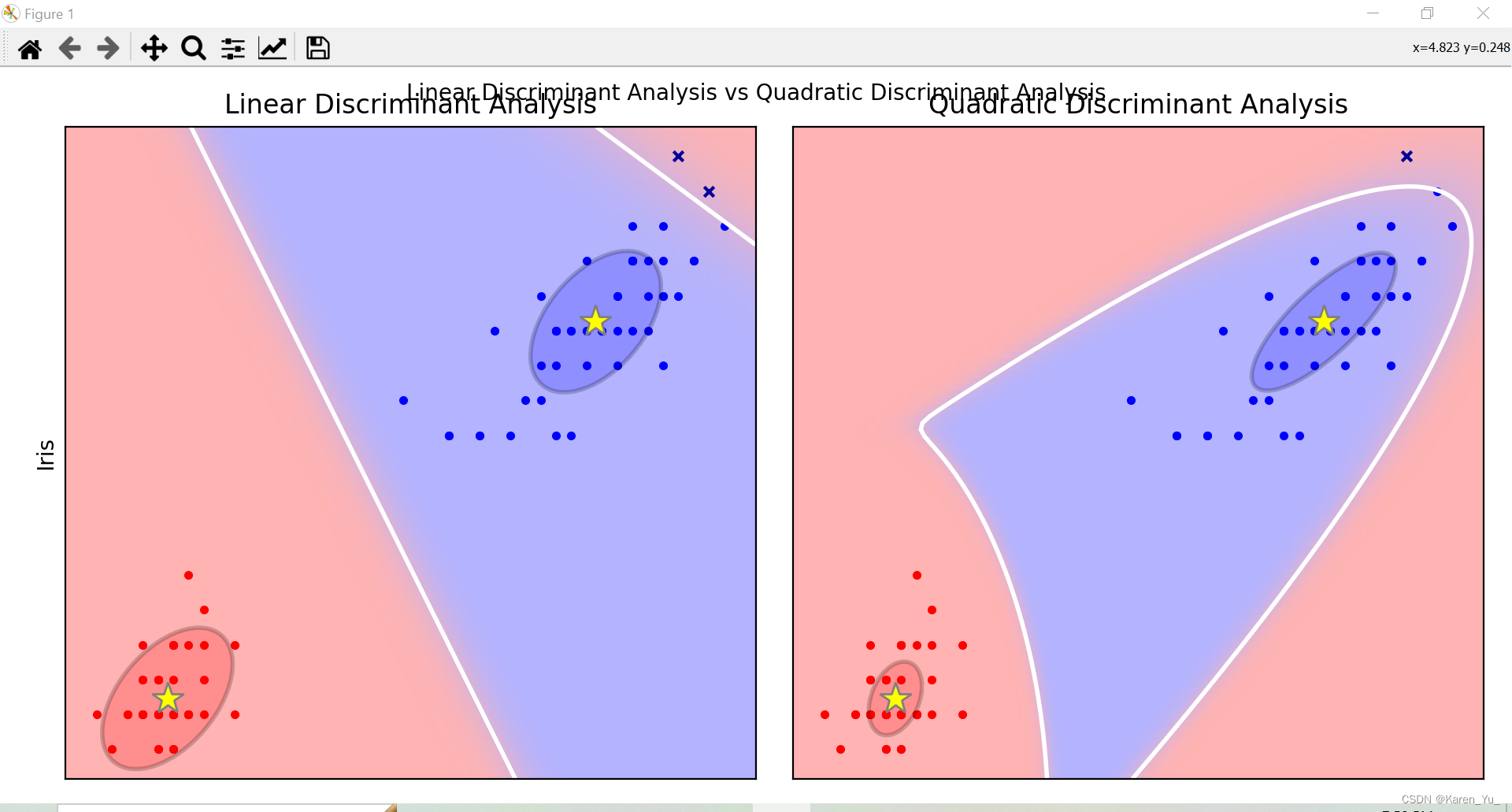
——————————————————————————————————————————
补充:今天看到了PCA的部分,想起来上学期看的autoencoder,遂又发生了一些谈话:
(前情提要,在说降维和特征选择的区别,一个是特征重组,一个是直接扔)
🐰:As I know, autoencoder can also combine features, for the encoder part, it is kind of similar to PCA?
💮:
Yes, that's correct. An autoencoder is a type of neural network that can be used for dimensionality reduction and feature extraction. Like PCA, it works by learning a compressed representation of the input data, but it does so in a nonlinear way.
The encoder part of an autoencoder is similar to PCA in that it projects the input data onto a lower-dimensional space, but it uses a neural network to learn a nonlinear mapping from the input space to the encoded space. This allows it to capture more complex patterns in the data than PCA.
The decoder part of the autoencoder maps the encoded data back to the original input space, and the network is trained to minimize the reconstruction error between the original input and the reconstructed output. By doing so, it learns a compressed representation of the input data that can be used for tasks such as data visualization, anomaly detection, or feature extraction.
One advantage of autoencoders over PCA is that they can learn more complex mappings between the input and encoded space, which can lead to better performance on some tasks. However, they can also be more difficult to train and may require more data than PCA to achieve good results.
🐰:Can I replace the encoder part in autoencoder by PCA, I know it might not get a good result, because PCA is not as complex as autoencoder.
💮:
It is possible to replace the encoder part of an autoencoder with PCA, but it is not recommended because it may not perform as well as using a neural network for the encoder.
One reason for this is that the encoder part of an autoencoder is specifically designed to learn a compressed representation of the input data that captures the most important patterns in the data. The neural network architecture used in the encoder is designed to learn a nonlinear mapping from the input space to the encoded space, which allows it to capture more complex patterns in the data than PCA. By using PCA instead of a neural network, you may not be able to capture all of the important patterns in the data, leading to a less effective compressed representation.
In addition, using PCA as the encoder would limit the flexibility of the autoencoder architecture, which could limit its ability to model complex data. One of the strengths of the autoencoder is its ability to learn more complex mappings between the input and encoded space than linear methods like PCA.
Overall, while it is possible to replace the encoder part of an autoencoder with PCA, it may not be the best approach in terms of performance and flexibility. It is generally recommended to use a neural network for the encoder part of an autoencoder to take advantage of its ability to learn complex mappings between the input and encoded space.
🐰:If I am building a neural network and I want to reduce the number of features, is it reasonable to use autoencoder rather than PCA or other dimentionality reduction method?
💮:
Using an autoencoder for feature reduction in a neural network can be a reasonable approach, especially if you have a large amount of data and want to capture more complex patterns in the data than linear methods like PCA can provide. Autoencoders are powerful models that can learn non-linear relationships between variables, making them a good choice for reducing the dimensionality of high-dimensional data.
However, it is important to keep in mind that using an autoencoder for feature reduction requires a lot of computational resources and training time. The training process can be time-consuming, and you may need to tune several hyperparameters to achieve good performance.
Another consideration is that autoencoders can be prone to overfitting if they are not regularized properly. This can be mitigated by using techniques like dropout, weight decay, or early stopping during the training process.
Overall, using an autoencoder for feature reduction in a neural network can be a powerful approach, but it should be done with care and attention to the computational and training requirements, as well as the risk of overfitting. Other dimensionality reduction methods like PCA or LDA may be faster and easier to implement, but they may not capture as complex patterns as autoencoders can.
🐰:Does that mean using dimensionality reduction methods will less likely cause overfitting than using autoencoder?
💮:
Using dimensionality reduction methods like PCA or LDA can help reduce the risk of overfitting compared to using autoencoders, especially if the number of retained features or components is small relative to the original number of features.
This is because dimensionality reduction methods are usually simpler and more transparent than neural networks, and they often impose more constraints on the learned representations to reduce the dimensionality of the input data. This can make it less likely that the reduced representations will overfit to the training data.
In contrast, autoencoders are more complex models that may be more prone to overfitting if not regularized properly. In addition, autoencoders may require a larger number of parameters to achieve good performance, which can increase the risk of overfitting if the model is too complex relative to the size of the training data.
However, it is important to keep in mind that overfitting can occur with any model if it is not properly regularized or validated. Therefore, it is always important to carefully tune and validate your models to ensure that they generalize well to new, unseen data.
忽然就串起来关于NN的一些回忆,复杂的模型确实能够更好的拟合训练的数据,但是也可能造成泛化能力不高(过拟合),简单的模型可能拟合训练数据做的不是特别好,但是在遇到新数据的时候可能反而表现的好,这在有关特征的处理上也是成立的。本质上,建立模型更像是trade off。
————————————————————————————————
有关autoencoder,简要来说就是一个压缩和解压缩,我们希望解压缩的结果和原始数据越接近越好(本质上,我们一定是希望能找到一个临界点,一旦跨过去,我们就丢失了重要的数据,以至于再也无法恢复原始数据,一旦没有走到临界点,那么我们的特征可能还是太多了)





















 8044
8044











 被折叠的 条评论
为什么被折叠?
被折叠的 条评论
为什么被折叠?








