5.使用lightgbm模型对新构造的特征进行模型训练和评估
前言
下面从数据探索,特征工程,模型训练,模型验证,特征优化,模型融合等六个方面完整分析工业蒸汽预测。参考内容为《阿里云天池大赛赛题解析-机器学习》书中内容。
一、业务理解
在用机器学习做业务分析时,一定要花些时间了解该业务所在的领域,该业务的流程,每个部分的重要点在哪里,数据具有什么的特点,常用的特征指标是什么,评估指标是什么,每个特征代表什么意思,要解决的问题是什么,分类,预测,聚类等等;只有充分了解该业务中数据的含义,才能在下面分析中构造或挑选合适的特征,从而构建适合的模型,进而更高效地解决问题。(一定要花些时间搞明白!)
在本例子中是做工业蒸汽分析,即要根据锅炉的工况预测产生的蒸汽量,训练集数据中有38个特征变量和target目标变量,数据是经过脱敏处理过的,为了保护数据隐私。测试集数据没有target字段,需要对它们进行预测。
很明显可以看出是回归预测问题,回归预测模型的预测结果是一个连续值域上的任意值,回归可以具有实值或离散的输入变量。同时输入变量按时间排序的回归问题称为时间序列预测问题。
回归预测模型常用算法包括线性回归,岭回归,LASSO回归,决策树回归,梯度提升树回归等,通常我们会用每种模型跑一下数据,从而从中选出最佳模型。
回归预测模型常用的评价指标均方误差。
二、数据探索
1.引入库
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
from scipy import stats
import warnings
warnings.filterwarnings("ignore")
%matplotlib inline
2.读入数据
使用Pandas库read_csv()函数进行数据提取,分割符为"\t"
train_data_file = "./zhengqi_train.txt"
test_data_file = "./zhengqi_test.txt"
train_data = pd.read_csv(train_data_file, sep='\t', encoding='utf-8')
test_data = pd.read_csv(test_data_file, sep='\t', encoding='utf-8')
3. 查看训练集测试集特征变量信息
train_data.info()
test_data.info()4.查看数据统计信息
train_data.describe()
test_data.describe()5. 画箱型图探索数据
# 画箱式图
column = train_data.columns.tolist()[:39] # 列表头
fig = plt.figure(figsize=(20, 40)) # 指定绘图对象宽度和高度
for i in range(38):
plt.subplot(13, 3, i + 1) # 13行3列子图
sns.boxplot(train_data[column[i]], orient="v", width=0.5) # 箱式图
plt.ylabel(column[i], fontsize=8)
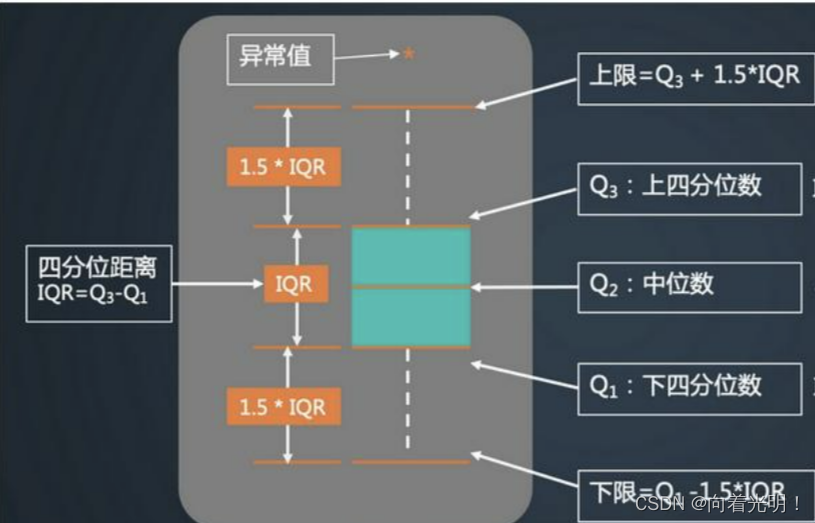
plt.show()箱型图以一种相对稳定的方式描述数据的离散分布情况,可以直观明了的识别数据批中的异常值。
6.查看数据分布图
查看查看所有数据的直方图和Q-Q图,查看训练集的数据是否近似于正态分布
train_cols = 6
train_rows = len(train_data.columns)
plt.figure(figsize=(4*train_cols,4*train_rows))
i=0
for col in train_data.columns:
i+=1
ax=plt.subplot(train_rows,train_cols,i)
sns.distplot(train_data[col],fit=stats.norm) #直方图
i+=1
ax=plt.subplot(train_rows,train_cols,i)
res = stats.probplot(train_data[col], plot=plt) # Q-Q图
plt.show()Q-Q图是指数据的分位数和正态分布的分位数对比参照的图,如果数据符合正态分布,则所有的点都会落在直线上。
直方图是一种统计报告图,它是根据具体数据的分布情况,画出以组距为底边,以频数为高度的一系列连接起来的直方型矩形图;用以展示数据的分布情况,诸如众数,中位数的大致位置,数据是否存在缺口或者异常值。
查看所有特征变量下,训练集数据和测试集数据的分布情况,分析并寻找出数据分布不一致的特征变量。
dist_cols = 6
dist_rows = len(test_data.columns)
plt.figure(figsize=(4*dist_cols,4*dist_rows))
i=1
for col in test_data.columns:
ax=plt.subplot(dist_rows,dist_cols,i)
ax = sns.kdeplot(train_data[col], color="Red", shade=True)
ax = sns.kdeplot(test_data[col], color="Blue", shade=True)
ax.set_xlabel(col)
ax.set_ylabel("Frequency")
ax = ax.legend(["train","test"])
i+=1
plt.show()KDE(Kernal Density Estimation,核密度估计)可以理解为是对直方图的加窗平滑,通过绘制KDE分布图,可以查看并对比训练集和测试集中特征变量的分布情况,发现两个数据集中分布不一致的特征变量。
训练集数据与测试集数据分布不一致,会导致模型泛化能力差,可采用删除此类特征的方法。
7.可视化线性回归关系
查看所有特征变量与'target'变量的线性回归关系
fcols = 6
frows = len(test_data.columns)
plt.figure(figsize=(5*fcols,4*frows))
i=0
for col in test_data.columns:
i+=1
ax=plt.subplot(frows,fcols,i)
sns.regplot(x=col, y='target', data=train_data, ax=ax,
scatter_kws={'marker':'.','s':3,'alpha':0.3},
line_kws={'color':'k'});
plt.xlabel(col)
plt.ylabel('target')
i+=1
ax=plt.subplot(frows,fcols,i)
sns.distplot(train_data[col].dropna())
plt.xlabel(col)查看特征变量的相关性
#这几个变量在训练集和测试集中数据分布一样
data_train1 = train_data.drop(['V5','V9','V11','V17','V22','V28'],axis=1)
train_corr = data_train1.corr()
train_corr
# 画出相关性热力图
ax = plt.subplots(figsize=(20, 16))#调整画布大小
ax = sns.heatmap(train_corr, vmax=.8, square=True, annot=True)#画热力图 annot=True 显示系数
# 找出相关程度
data_train1 = train_data.drop(['V5','V9','V11','V17','V22','V28'],axis=1)
plt.figure(figsize=(20, 16)) # 指定绘图对象宽度和高度
colnm = data_train1.columns.tolist() # 列表头
mcorr = data_train1[colnm].corr(method="spearman") # 相关系数矩阵,即给出了任意两个变量之间的相关系数
mask = np.zeros_like(mcorr, dtype=np.bool) # 构造与mcorr同维数矩阵 为bool型
mask[np.triu_indices_from(mask)] = True # 角分线右侧为True
cmap = sns.diverging_palette(220, 10, as_cmap=True) # 返回matplotlib colormap对象
g = sns.heatmap(mcorr, mask=mask, cmap=cmap, square=True, annot=True, fmt='0.2f') # 热力图(看两两相似度)
plt.show()查找出特征变量和target变量相关系数大于0.5的特征变量
#寻找K个最相关的特征信息
k = 10 # number of variables for heatmap
cols = train_corr.nlargest(k, 'target')['target'].index
cm = np.corrcoef(train_data[cols].values.T)
hm = plt.subplots(figsize=(10, 10))#调整画布大小
#hm = sns.heatmap(cm, cbar=True, annot=True, square=True)
#g = sns.heatmap(train_data[cols].corr(),annot=True,square=True,cmap="RdYlGn")
hm = sns.heatmap(train_data[cols].corr(),annot=True,square=True)
plt.show()
threshold = 0.5
corrmat = train_data.corr()
top_corr_features = corrmat.index[abs(corrmat["target"])>threshold]
plt.figure(figsize=(10,10))
g = sns.heatmap(train_data[top_corr_features].corr(),annot=True,cmap="RdYlGn")
相关性选择主要用于判别线性相关,对于target变量如果存在更复杂的函数形式的影响,则建议使用树模型的特征重要性去选择。
用相关系数阈值移除相关特征
# Threshold for removing correlated variables
threshold = 0.5
# Absolute value correlation matrix
corr_matrix = data_train1.corr().abs()
drop_col=corr_matrix[corr_matrix["target"]<threshold].index
#data_all.drop(drop_col, axis=1, inplace=True)Box-Cox变换
由于线性回归是基于正态分布的,因此在进行统计分析时,需要将数据转换使其符合正态分布。Box-Cox变换是统计建模中常用的一种数据转换方法。可以使线性回归模型在满足线性,正态性,独立性及方差齐性的同时,又不丢失信息。在一定程度上减小不可预测的误差和预测变量的相关性,有利于线性模型的拟合及分析出特征的相关性。
在做Box-Cox变换之前,需要对数据做归一化预处理。
#合并训练集和测试集数据
train_x = train_data.drop(['target'], axis=1)
#data_all=pd.concat([train_data,test_data],axis=0,ignore_index=True)
data_all = pd.concat([train_x,test_data])
data_all.drop(drop_columns,axis=1,inplace=True)
#View data
data_all.head()
# 对合并后的每列数据进行归一化
cols_numeric=list(data_all.columns)
def scale_minmax(col):
return (col-col.min())/(col.max()-col.min())
data_all[cols_numeric] = data_all[cols_numeric].apply(scale_minmax,axis=0)
data_all[cols_numeric].describe()
#分开对训练集和测试集数据进行归一化处理
#col_data_process = cols_numeric.append('target')
train_data_process = train_data[cols_numeric]
train_data_process = train_data_process[cols_numeric].apply(scale_minmax,axis=0)
test_data_process = test_data[cols_numeric]
test_data_process = test_data_process[cols_numeric].apply(scale_minmax,axis=0)
cols_numeric_left = cols_numeric[0:13]
cols_numeric_right = cols_numeric[13:]
## Check effect of Box-Cox transforms on distributions of continuous variables
train_data_process = pd.concat([train_data_process, train_data['target']], axis=1)
fcols = 6
frows = len(cols_numeric_left)
plt.figure(figsize=(4*fcols,4*frows))
i=0
for var in cols_numeric_left:
dat = train_data_process[[var, 'target']].dropna()
i+=1
plt.subplot(frows,fcols,i)
sns.distplot(dat[var] , fit=stats.norm);
plt.title(var+' Original')
plt.xlabel('')
i+=1
plt.subplot(frows,fcols,i)
_=stats.probplot(dat[var], plot=plt)
plt.title('skew='+'{:.4f}'.format(stats.skew(dat[var])))
plt.xlabel('')
plt.ylabel('')
i+=1
plt.subplot(frows,fcols,i)
plt.plot(dat[var], dat['target'],'.',alpha=0.5)
plt.title('corr='+'{:.2f}'.format(np.corrcoef(dat[var], dat['target'])[0][1]))
i+=1
plt.subplot(frows,fcols,i)
trans_var, lambda_var = stats.boxcox(dat[var].dropna()+1)
trans_var = scale_minmax(trans_var)
sns.distplot(trans_var , fit=stats.norm);
plt.title(var+' Tramsformed')
plt.xlabel('')
i+=1
plt.subplot(frows,fcols,i)
_=stats.probplot(trans_var, plot=plt)
plt.title('skew='+'{:.4f}'.format(stats.skew(trans_var)))
plt.xlabel('')
plt.ylabel('')
i+=1
plt.subplot(frows,fcols,i)
plt.plot(trans_var, dat['target'],'.',alpha=0.5)
plt.title('corr='+'{:.2f}'.format(np.corrcoef(trans_var,dat['target'])[0][1]))三、特征工程
特征工程就是从原始数据提取特征的过程,这些特征可以很好地描述数据,特征工程一般包括特征使用,特征获取,特征处理,特征选择和特征监控等。
1.导入数据分析工具包
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
from scipy import stats
import warnings
warnings.filterwarnings("ignore")
%matplotlib inline2.数据读取
train_data_file = "./zhengqi_train.txt"
test_data_file = "./zhengqi_test.txt"
train_data = pd.read_csv(train_data_file, sep='\t', encoding='utf-8')
test_data = pd.read_csv(test_data_file, sep='\t', encoding='utf-8')3.异常值分析
plt.figure(figsize=(18, 10))
plt.boxplot(x=train_data.values,labels=train_data.columns)
plt.hlines([-7.5, 7.5], 0, 40, colors='r')
plt.show()对于异常值进行删除
4.最大值最小值归一化
from sklearn import preprocessing
features_columns = [col for col in train_data.columns if col not in ['target']]
min_max_scaler = preprocessing.MinMaxScaler()
min_max_scaler = min_max_scaler.fit(train_data[features_columns])
train_data_scaler = min_max_scaler.transform(train_data[features_columns])
test_data_scaler = min_max_scaler.transform(test_data[features_columns])
train_data_scaler = pd.DataFrame(train_data_scaler)
train_data_scaler.columns = features_columns
test_data_scaler = pd.DataFrame(test_data_scaler)
test_data_scaler.columns = features_columns
train_data_scaler['target'] = train_data['target']5.查看训练集数据和测试集数据分布情况
dist_cols = 6
dist_rows = len(test_data_scaler.columns)
plt.figure(figsize=(4*dist_cols,4*dist_rows))
for i, col in enumerate(test_data_scaler.columns):
ax=plt.subplot(dist_rows,dist_cols,i+1)
ax = sns.kdeplot(train_data_scaler[col], color="Red", shade=True)
ax = sns.kdeplot(test_data_scaler[col], color="Blue", shade=True)
ax.set_xlabel(col)
ax.set_ylabel("Frequency")
ax = ax.legend(["train","test"])
plt.show()6. 特征相关性
plt.figure(figsize=(20, 16))
column = train_data_scaler.columns.tolist()
mcorr = train_data_scaler[column].corr(method="spearman")
mask = np.zeros_like(mcorr, dtype=np.bool)
mask[np.triu_indices_from(mask)] = True
cmap = sns.diverging_palette(220, 10, as_cmap=True)
g = sns.heatmap(mcorr, mask=mask, cmap=cmap, square=True, annot=True, fmt='0.2f')
plt.show()7.特征降维
相关性分析
mcorr=mcorr.abs()
numerical_corr=mcorr[mcorr['target']>0.1]['target']
print(numerical_corr.sort_values(ascending=False))
index0 = numerical_corr.sort_values(ascending=False).index
print(train_data_scaler[index0].corr('spearman'))相关性初筛
features_corr = numerical_corr.sort_values(ascending=False).reset_index()
features_corr.columns = ['features_and_target', 'corr']
features_corr_select = features_corr[features_corr['corr']>0.3] # 筛选出大于相关性大于0.3的特征
print(features_corr_select)
select_features = [col for col in features_corr_select['features_and_target'] if col not in ['target']]
new_train_data_corr_select = train_data_scaler[select_features+['target']]
new_test_data_corr_select = test_data_scaler[select_features]多重共线性分析
from statsmodels.stats.outliers_influence import variance_inflation_factor #多重共线性方差膨胀因子
#多重共线性
new_numerical=['V0', 'V2', 'V3', 'V4', 'V5', 'V6', 'V10','V11',
'V13', 'V15', 'V16', 'V18', 'V19', 'V20', 'V22','V24','V30', 'V31', 'V37']
X=np.matrix(train_data_scaler[new_numerical])
VIF_list=[variance_inflation_factor(X, i) for i in range(X.shape[1])]
VIF_listPCA去除多重共线性 降维
from sklearn.decomposition import PCA #主成分分析法
#PCA方法降维
#保持90%的信息
pca = PCA(n_components=0.9)
new_train_pca_90 = pca.fit_transform(train_data_scaler.iloc[:,0:-1])
new_test_pca_90 = pca.transform(test_data_scaler)
new_train_pca_90 = pd.DataFrame(new_train_pca_90)
new_test_pca_90 = pd.DataFrame(new_test_pca_90)
new_train_pca_90['target'] = train_data_scaler['target']
new_train_pca_90.describe()#PCA方法降维
#保留16个主成分
pca = PCA(n_components=0.95)
new_train_pca_16 = pca.fit_transform(train_data_scaler.iloc[:,0:-1])
new_test_pca_16 = pca.transform(test_data_scaler)
new_train_pca_16 = pd.DataFrame(new_train_pca_16)
new_test_pca_16 = pd.DataFrame(new_test_pca_16)
new_train_pca_16['target'] = train_data_scaler['target']
new_train_pca_16.describe()四、模型训练
1.导入相关库
from sklearn.linear_model import LinearRegression #线性回归
from sklearn.neighbors import KNeighborsRegressor #K近邻回归
from sklearn.tree import DecisionTreeRegressor #决策树回归
from sklearn.ensemble import RandomForestRegressor #随机森林回归
from sklearn.svm import SVR #支持向量回归
import lightgbm as lgb #lightGbm模型
from sklearn.ensemble import GradientBoostingRegressor
from sklearn.model_selection import train_test_split # 切分数据
from sklearn.metrics import mean_squared_error #评价指标2.切分训练数据和线下验证数据
#采用 pca 保留16维特征的数据
new_train_pca_16 = new_train_pca_16.fillna(0)
train = new_train_pca_16[new_test_pca_16.columns]
target = new_train_pca_16['target']
# 切分数据 训练数据80% 验证数据20%
train_data,test_data,train_target,test_target=train_test_split(train,target,test_size=0.2,random_state=0)3.定义绘制学习曲线函数
def plot_learning_curve(estimator, title, X, y, ylim=None, cv=None,
n_jobs=1, train_sizes=np.linspace(.1, 1.0, 5)):
plt.figure()
plt.title(title)
if ylim is not None:
plt.ylim(*ylim)
plt.xlabel("Training examples")
plt.ylabel("Score")
train_sizes, train_scores, test_scores = learning_curve(
estimator, X, y, cv=cv, n_jobs=n_jobs, train_sizes=train_sizes)
train_scores_mean = np.mean(train_scores, axis=1)
train_scores_std = np.std(train_scores, axis=1)
test_scores_mean = np.mean(test_scores, axis=1)
test_scores_std = np.std(test_scores, axis=1)
print(train_scores_mean)
print(test_scores_mean)
plt.grid()
plt.fill_between(train_sizes, train_scores_mean - train_scores_std,
train_scores_mean + train_scores_std, alpha=0.1,
color="r")
plt.fill_between(train_sizes, test_scores_mean - test_scores_std,
test_scores_mean + test_scores_std, alpha=0.1, color="g")
plt.plot(train_sizes, train_scores_mean, 'o-', color="r",
label="Training score")
plt.plot(train_sizes, test_scores_mean, 'o-', color="g",
label="Cross-validation score")
plt.legend(loc="best")
return plt4.多元线性回归模型
clf = LinearRegression()
clf.fit(train_data, train_target)
score = mean_squared_error(test_target, clf.predict(test_data))
print("LinearRegression: ", score)5.绘制线性回归模型学习曲线
X = train_data.values
y = train_target.values
title = r"LinearRegression"
cv = ShuffleSplit(n_splits=100, test_size=0.2, random_state=0)
estimator = LinearRegression() #建模
plot_learning_curve(estimator, title, X, y, ylim=(0.5, 0.8), cv=cv, n_jobs=1)6.K近邻回归
clf = KNeighborsRegressor(n_neighbors=8) # 最近三个
clf.fit(train_data, train_target)
score = mean_squared_error(test_target, clf.predict(test_data))
print("KNeighborsRegressor: ", score)7.绘制K近邻回归学习曲线
X = train_data.values
y = train_target.values
# K近邻回归
title = r"KNeighborsRegressor"
cv = ShuffleSplit(n_splits=100, test_size=0.2, random_state=0)
estimator = KNeighborsRegressor(n_neighbors=8) #建模
plot_learning_curve(estimator, title, X, y, ylim=(0.3, 0.9), cv=cv, n_jobs=1)8.决策树回归
clf.fit(train_data, train_target)
score = mean_squared_error(test_target, clf.predict(test_data))
print("DecisionTreeRegressor: ", score)9.绘制决策树学习曲线
X = train_data.values
y = train_target.values
# 决策树回归
title = r"DecisionTreeRegressor"
cv = ShuffleSplit(n_splits=100, test_size=0.2, random_state=0)
estimator = DecisionTreeRegressor() #建模
plot_learning_curve(estimator, title, X, y, ylim=(0.1, 1.3), cv=cv, n_jobs=1)10.随机森林回归
clf = RandomForestRegressor(n_estimators=200) # 200棵树模型
clf.fit(train_data, train_target)
score = mean_squared_error(test_target, clf.predict(test_data))
print("RandomForestRegressor: ", score)11.绘制随机森林回归学习曲线
X = train_data.values
y = train_target.values
# K近邻回归
title = r"RandomForestRegressor"
cv = ShuffleSplit(n_splits=100, test_size=0.2, random_state=0)
estimator = RandomForestRegressor(n_estimators=200) #建模
plot_learning_curve(estimator, title, X, y, ylim=(0.4, 1.0), cv=cv, n_jobs=1)12.Lgb模型回归
# lgb回归模型
clf = lgb.LGBMRegressor(
learning_rate=0.01,
max_depth=-1,
n_estimators=5000,
boosting_type='gbdt',
random_state=2019,
objective='regression',
)
# 训练模型
clf.fit(
X=train_data, y=train_target,
eval_metric='MSE',
verbose=50
)
score = mean_squared_error(test_target, clf.predict(test_data))
print("lightGbm: ", score)13.绘制lgb回归学习曲线
X = train_data.values
y = train_target.values
# K近邻回归
title = r"LGBMRegressor"
cv = ShuffleSplit(n_splits=100, test_size=0.2, random_state=0)
estimator = lgb.LGBMRegressor(
learning_rate=0.01,
max_depth=-1,
n_estimators=100,
boosting_type='gbdt',
random_state=2019,
objective='regression'
) #建模
plot_learning_curve(estimator, title, X, y, ylim=(0.4, 1.0), cv=cv, n_jobs=1)14.绘制学习曲线
def plot_learning_curve(algo, X_train, X_test, y_train, y_test):
"""绘制学习曲线:只需要传入算法(或实例对象)、X_train、X_test、y_train、y_test"""
"""当使用该函数时传入算法,该算法的变量要进行实例化,如:PolynomialRegression(degree=2),变量 degree 要进行实例化"""
train_score = []
test_score = []
for i in range(10, len(X_train)+1, 10):
algo.fit(X_train[:i], y_train[:i])
y_train_predict = algo.predict(X_train[:i])
train_score.append(mean_squared_error(y_train[:i], y_train_predict))
y_test_predict = algo.predict(X_test)
test_score.append(mean_squared_error(y_test, y_test_predict))
plt.plot([i for i in range(1, len(train_score)+1)],
train_score, label="train")
plt.plot([i for i in range(1, len(test_score)+1)],
test_score, label="test")
plt.legend()
plt.show()五、模型验证
1.模型过拟合与欠拟合
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
from scipy import stats
import warnings
warnings.filterwarnings("ignore")
from sklearn.linear_model import LinearRegression #线性回归
from sklearn.neighbors import KNeighborsRegressor #K近邻回归
from sklearn.tree import DecisionTreeRegressor #决策树回归
from sklearn.ensemble import RandomForestRegressor #随机森林回归
from sklearn.svm import SVR #支持向量回归
import lightgbm as lgb #lightGbm模型
from sklearn.model_selection import train_test_split # 切分数据
from sklearn.metrics import mean_squared_error #评价指标
from sklearn.linear_model import SGDRegressortrain_data_file = "./zhengqi_train.txt"
test_data_file = "./zhengqi_test.txt"
train_data = pd.read_csv(train_data_file, sep='\t', encoding='utf-8')
test_data = pd.read_csv(test_data_file, sep='\t', encoding='utf-8')from sklearn import preprocessing
features_columns = [col for col in train_data.columns if col not in ['target']]
min_max_scaler = preprocessing.MinMaxScaler()
min_max_scaler = min_max_scaler.fit(train_data[features_columns])
train_data_scaler = min_max_scaler.transform(train_data[features_columns])
test_data_scaler = min_max_scaler.transform(test_data[features_columns])
train_data_scaler = pd.DataFrame(train_data_scaler)
train_data_scaler.columns = features_columns
test_data_scaler = pd.DataFrame(test_data_scaler)
test_data_scaler.columns = features_columns
train_data_scaler['target'] = train_data['target']from sklearn.decomposition import PCA #主成分分析法
#PCA方法降维
#保留16个主成分
pca = PCA(n_components=16)
new_train_pca_16 = pca.fit_transform(train_data_scaler.iloc[:,0:-1])
new_test_pca_16 = pca.transform(test_data_scaler)
new_train_pca_16 = pd.DataFrame(new_train_pca_16)
new_test_pca_16 = pd.DataFrame(new_test_pca_16)
new_train_pca_16['target'] = train_data_scaler['target']#采用 pca 保留16维特征的数据
new_train_pca_16 = new_train_pca_16.fillna(0)
train = new_train_pca_16[new_test_pca_16.columns]
target = new_train_pca_16['target']
# 切分数据 训练数据80% 验证数据20%
train_data,test_data,train_target,test_target=train_test_split(train,target,test_size=0.2,random_state=0)欠拟合
clf = SGDRegressor(max_iter=500, tol=1e-2)
clf.fit(train_data, train_target)
score_train = mean_squared_error(train_target, clf.predict(train_data))
score_test = mean_squared_error(test_target, clf.predict(test_data))
print("SGDRegressor train MSE: ", score_train)
print("SGDRegressor test MSE: ", score_test)过拟合
from sklearn.preprocessing import PolynomialFeatures
poly = PolynomialFeatures(5)
train_data_poly = poly.fit_transform(train_data)
test_data_poly = poly.transform(test_data)
clf = SGDRegressor(max_iter=1000, tol=1e-3)
clf.fit(train_data_poly, train_target)
score_train = mean_squared_error(train_target, clf.predict(train_data_poly))
score_test = mean_squared_error(test_target, clf.predict(test_data_poly))
print("SGDRegressor train MSE: ", score_train)
print("SGDRegressor test MSE: ", score_test)正常拟合
from sklearn.preprocessing import PolynomialFeatures
poly = PolynomialFeatures(3)
train_data_poly = poly.fit_transform(train_data)
test_data_poly = poly.transform(test_data)
clf = SGDRegressor(max_iter=1000, tol=1e-3)
clf.fit(train_data_poly, train_target)
score_train = mean_squared_error(train_target, clf.predict(train_data_poly))
score_test = mean_squared_error(test_target, clf.predict(test_data_poly))
print("SGDRegressor train MSE: ", score_train)
print("SGDRegressor test MSE: ", score_test)2.模型正则化
L2范数正则化
poly = PolynomialFeatures(3)
train_data_poly = poly.fit_transform(train_data)
test_data_poly = poly.transform(test_data)
clf = SGDRegressor(max_iter=1000, tol=1e-3, penalty= 'L2', alpha=0.0001)
clf.fit(train_data_poly, train_target)
score_train = mean_squared_error(train_target, clf.predict(train_data_poly))
score_test = mean_squared_error(test_target, clf.predict(test_data_poly))
print("SGDRegressor train MSE: ", score_train)
print("SGDRegressor test MSE: ", score_test)L1范数正则化
poly = PolynomialFeatures(3)
train_data_poly = poly.fit_transform(train_data)
test_data_poly = poly.transform(test_data)
clf = SGDRegressor(max_iter=1000, tol=1e-3, penalty= 'L1', alpha=0.00001)
clf.fit(train_data_poly, train_target)
score_train = mean_squared_error(train_target, clf.predict(train_data_poly))
score_test = mean_squared_error(test_target, clf.predict(test_data_poly))
print("SGDRegressor train MSE: ", score_train)
print("SGDRegressor test MSE: ", score_test)ElasticNet L1和L2范数加权正则化
poly = PolynomialFeatures(3)
train_data_poly = poly.fit_transform(train_data)
test_data_poly = poly.transform(test_data)
clf = SGDRegressor(max_iter=1000, tol=1e-3, penalty= 'elasticnet', l1_ratio=0.9, alpha=0.00001)
clf.fit(train_data_poly, train_target)
score_train = mean_squared_error(train_target, clf.predict(train_data_poly))
score_test = mean_squared_error(test_target, clf.predict(test_data_poly))
print("SGDRegressor train MSE: ", score_train)
print("SGDRegressor test MSE: ", score_test)3.模型交叉验证
简单交叉验证
# 简单交叉验证
from sklearn.model_selection import train_test_split # 切分数据
# 切分数据 训练数据80% 验证数据20%
train_data,test_data,train_target,test_target=train_test_split(train,target,test_size=0.2,random_state=0)
clf = SGDRegressor(max_iter=1000, tol=1e-3)
clf.fit(train_data, train_target)
score_train = mean_squared_error(train_target, clf.predict(train_data))
score_test = mean_squared_error(test_target, clf.predict(test_data))
print("SGDRegressor train MSE: ", score_train)
print("SGDRegressor test MSE: ", score_test)K折交叉验证K-fold CV
# 5折交叉验证
from sklearn.model_selection import KFold
kf = KFold(n_splits=5)
for k, (train_index, test_index) in enumerate(kf.split(train)):
train_data,test_data,train_target,test_target = train.values[train_index],train.values[test_index],target[train_index],target[test_index]
clf = SGDRegressor(max_iter=1000, tol=1e-3)
clf.fit(train_data, train_target)
score_train = mean_squared_error(train_target, clf.predict(train_data))
score_test = mean_squared_error(test_target, clf.predict(test_data))
print(k, " 折", "SGDRegressor train MSE: ", score_train)
print(k, " 折", "SGDRegressor test MSE: ", score_test, '\n') 留一法 LOO CV
from sklearn.model_selection import LeaveOneOut
loo = LeaveOneOut()
num = 100
for k, (train_index, test_index) in enumerate(loo.split(train)):
train_data,test_data,train_target,test_target = train.values[train_index],train.values[test_index],target[train_index],target[test_index]
clf = SGDRegressor(max_iter=1000, tol=1e-3)
clf.fit(train_data, train_target)
score_train = mean_squared_error(train_target, clf.predict(train_data))
score_test = mean_squared_error(test_target, clf.predict(test_data))
print(k, " 个", "SGDRegressor train MSE: ", score_train)
print(k, " 个", "SGDRegressor test MSE: ", score_test, '\n')
if k >= 9:
break留P法 LPO CV
from sklearn.model_selection import LeavePOut
lpo = LeavePOut(p=10)
num = 100
for k, (train_index, test_index) in enumerate(lpo.split(train)):
train_data,test_data,train_target,test_target = train.values[train_index],train.values[test_index],target[train_index],target[test_index]
clf = SGDRegressor(max_iter=1000, tol=1e-3)
clf.fit(train_data, train_target)
score_train = mean_squared_error(train_target, clf.predict(train_data))
score_test = mean_squared_error(test_target, clf.predict(test_data))
print(k, " 10个", "SGDRegressor train MSE: ", score_train)
print(k, " 10个", "SGDRegressor test MSE: ", score_test, '\n')
if k >= 9:
break4.模型超参空间及调参
穷举网格搜索
from sklearn.model_selection import GridSearchCV
from sklearn.ensemble import RandomForestRegressor
from sklearn.model_selection import train_test_split # 切分数据
# 切分数据 训练数据80% 验证数据20%
train_data,test_data,train_target,test_target=train_test_split(train,target,test_size=0.2,random_state=0)
randomForestRegressor = RandomForestRegressor()
parameters = {
'n_estimators':[50, 100, 200],
'max_depth':[1, 2, 3]
}
clf = GridSearchCV(randomForestRegressor, parameters, cv=5)
clf.fit(train_data, train_target)
score_test = mean_squared_error(test_target, clf.predict(test_data))
print("RandomForestRegressor GridSearchCV test MSE: ", score_test)
sorted(clf.cv_results_.keys())随机参数优化
from sklearn.model_selection import RandomizedSearchCV
from sklearn.ensemble import RandomForestRegressor
from sklearn.model_selection import train_test_split # 切分数据
# 切分数据 训练数据80% 验证数据20%
train_data,test_data,train_target,test_target=train_test_split(train,target,test_size=0.2,random_state=0)
randomForestRegressor = RandomForestRegressor()
parameters = {
'n_estimators':[50, 100, 200, 300],
'max_depth':[1, 2, 3, 4, 5]
}
clf = RandomizedSearchCV(randomForestRegressor, parameters, cv=5)
clf.fit(train_data, train_target)
score_test = mean_squared_error(test_target, clf.predict(test_data))
print("RandomForestRegressor RandomizedSearchCV test MSE: ", score_test)
sorted(clf.cv_results_.keys())Lgb调参
clf = lgb.LGBMRegressor(num_leaves=31)
parameters = {
'learning_rate': [0.01, 0.1, 1],
'n_estimators': [20, 40]
}
clf = GridSearchCV(clf, parameters, cv=5)
clf.fit(train_data, train_target)
print('Best parameters found by grid search are:', clf.best_params_)
score_test = mean_squared_error(test_target, clf.predict(test_data))
print("LGBMRegressor RandomizedSearchCV test MSE: ", score_test)Lgb线下验证
train_data2 = pd.read_csv('./zhengqi_train.txt',sep='\t')
test_data2 = pd.read_csv('./zhengqi_test.txt',sep='\t')
train_data2_f = train_data2[test_data2.columns].values
train_data2_target = train_data2['target'].values
# lgb 模型
from sklearn.model_selection import KFold
import lightgbm as lgb
import numpy as np
# 5折交叉验证
Folds=5
kf = KFold(len(train_data2_f), n_splits=Folds, random_state=100, shuffle=True)
# 记录训练和预测MSE
MSE_DICT = {
'train_mse':[],
'test_mse':[]
}
# 线下训练预测
for i, (train_index, test_index) in enumerate(kf.split(train_data2_f)):
# lgb树模型
lgb_reg = lgb.LGBMRegressor(
learning_rate=0.01,
max_depth=-1,
n_estimators=100,
boosting_type='gbdt',
random_state=100,
objective='regression',
)
# 切分训练集和预测集
X_train_KFold, X_test_KFold = train_data2_f[train_index], train_data2_f[test_index]
y_train_KFold, y_test_KFold = train_data2_target[train_index], train_data2_target[test_index]
# 训练模型
# reg.fit(X_train_KFold, y_train_KFold)
lgb_reg.fit(
X=X_train_KFold,y=y_train_KFold,
eval_set=[(X_train_KFold, y_train_KFold),(X_test_KFold, y_test_KFold)],
eval_names=['Train','Test'],
early_stopping_rounds=100,
eval_metric='MSE',
verbose=50
)
# 训练集预测 测试集预测
y_train_KFold_predict = lgb_reg.predict(X_train_KFold,num_iteration=lgb_reg.best_iteration_)
y_test_KFold_predict = lgb_reg.predict(X_test_KFold,num_iteration=lgb_reg.best_iteration_)
print('第{}折 训练和预测 训练MSE 预测MSE'.format(i))
train_mse = mean_squared_error(y_train_KFold_predict, y_train_KFold)
print('------\n', '训练MSE\n', train_mse, '\n------')
test_mse = mean_squared_error(y_test_KFold_predict, y_test_KFold)
print('------\n', '预测MSE\n', test_mse, '\n------\n')
MSE_DICT['train_mse'].append(train_mse)
MSE_DICT['test_mse'].append(test_mse)
print('------\n', '训练MSE\n', MSE_DICT['train_mse'], '\n', np.mean(MSE_DICT['train_mse']), '\n------')
print('------\n', '预测MSE\n', MSE_DICT['test_mse'], '\n', np.mean(MSE_DICT['test_mse']), '\n------')5.学习曲线和验证曲线
学习曲线
import numpy as np
import matplotlib.pyplot as plt
from sklearn import model_selection
from sklearn.linear_model import SGDRegressor
from sklearn.model_selection import learning_curve
def plot_learning_curve(estimator, title, X, y, ylim=None, cv=None,
n_jobs=1, train_sizes=np.linspace(.1, 1.0, 5)):
plt.figure()
plt.title(title)
if ylim is not None:
plt.ylim(*ylim)
plt.xlabel("Training examples")
plt.ylabel("Score")
train_sizes, train_scores, test_scores = learning_curve(
estimator, X, y, cv=cv, n_jobs=n_jobs, train_sizes=train_sizes)
train_scores_mean = np.mean(train_scores, axis=1)
train_scores_std = np.std(train_scores, axis=1)
test_scores_mean = np.mean(test_scores, axis=1)
test_scores_std = np.std(test_scores, axis=1)
plt.grid()
plt.fill_between(train_sizes, train_scores_mean - train_scores_std,
train_scores_mean + train_scores_std, alpha=0.1,
color="r")
plt.fill_between(train_sizes, test_scores_mean - test_scores_std,
test_scores_mean + test_scores_std, alpha=0.1, color="g")
plt.plot(train_sizes, train_scores_mean, 'o-', color="r",
label="Training score")
plt.plot(train_sizes, test_scores_mean, 'o-', color="g",
label="Cross-validation score")
plt.legend(loc="best")
return plt
X = train_data2[test_data2.columns].values
y = train_data2['target'].values
title = "LinearRegression"
# Cross validation with 100 iterations to get smoother mean test and train
# score curves, each time with 20% data randomly selected as a validation set.
cv = model_selection.ShuffleSplit(X.shape[0], n_splits=100,
test_size=0.2, random_state=0)
estimator = SGDRegressor()
plot_learning_curve(estimator, title, X, y, ylim=(0.7, 1.01), cv=cv, n_jobs=-1)验证曲线
import matplotlib.pyplot as plt
import numpy as np
from sklearn.linear_model import SGDRegressor
from sklearn.model_selection import validation_curve
X = train_data2[test_data2.columns].values
y = train_data2['target'].values
# max_iter=1000, tol=1e-3, penalty= 'L1', alpha=0.00001
param_range = [0.1, 0.01, 0.001, 0.0001, 0.00001, 0.000001]
train_scores, test_scores = validation_curve(
SGDRegressor(max_iter=1000, tol=1e-3, penalty= 'L1'), X, y, param_name="alpha", param_range=param_range,
cv=10, scoring='r2', n_jobs=1)
train_scores_mean = np.mean(train_scores, axis=1)
train_scores_std = np.std(train_scores, axis=1)
test_scores_mean = np.mean(test_scores, axis=1)
test_scores_std = np.std(test_scores, axis=1)
plt.title("Validation Curve with SGDRegressor")
plt.xlabel("alpha")
plt.ylabel("Score")
plt.ylim(0.0, 1.1)
plt.semilogx(param_range, train_scores_mean, label="Training score", color="r")
plt.fill_between(param_range, train_scores_mean - train_scores_std,
train_scores_mean + train_scores_std, alpha=0.2, color="r")
plt.semilogx(param_range, test_scores_mean, label="Cross-validation score",
color="g")
plt.fill_between(param_range, test_scores_mean - test_scores_std,
test_scores_mean + test_scores_std, alpha=0.2, color="g")
plt.legend(loc="best")
plt.show()六、特征优化
1.导入数据
import pandas as pd
train_data_file = "./zhengqi_train.txt"
test_data_file = "./zhengqi_test.txt"
train_data = pd.read_csv(train_data_file, sep='\t', encoding='utf-8')
test_data = pd.read_csv(test_data_file, sep='\t', encoding='utf-8')2.定义特征构造方法,构造特征
epsilon=1e-5
#组交叉特征,可以自行定义,如增加: x*x/y, log(x)/y 等等
func_dict = {
'add': lambda x,y: x+y,
'mins': lambda x,y: x-y,
'div': lambda x,y: x/(y+epsilon),
'multi': lambda x,y: x*y
}3.定义特征构造的函数
def auto_features_make(train_data,test_data,func_dict,col_list):
train_data, test_data = train_data.copy(), test_data.copy()
for col_i in col_list:
for col_j in col_list:
for func_name, func in func_dict.items():
for data in [train_data,test_data]:
func_features = func(data[col_i],data[col_j])
col_func_features = '-'.join([col_i,func_name,col_j])
data[col_func_features] = func_features
return train_data,test_data4.对训练集和测试集数据进行特征构造
train_data2, test_data2 = auto_features_make(train_data,test_data,func_dict,col_list=test_data.columns)from sklearn.decomposition import PCA #主成分分析法
#PCA方法降维
pca = PCA(n_components=500)
train_data2_pca = pca.fit_transform(train_data2.iloc[:,0:-1])
test_data2_pca = pca.transform(test_data2)
train_data2_pca = pd.DataFrame(train_data2_pca)
test_data2_pca = pd.DataFrame(test_data2_pca)
train_data2_pca['target'] = train_data2['target']X_train2 = train_data2[test_data2.columns].values
y_train = train_data2['target']5.使用lightgbm模型对新构造的特征进行模型训练和评估
# ls_validation i
from sklearn.model_selection import KFold
from sklearn.metrics import mean_squared_error
import lightgbm as lgb
import numpy as np
# 5折交叉验证
Folds=5
kf = KFold(len(X_train2), n_splits=Folds, random_state=2019, shuffle=True)
# 记录训练和预测MSE
MSE_DICT = {
'train_mse':[],
'test_mse':[]
}
# 线下训练预测
for i, (train_index, test_index) in enumerate(kf.split(X_train2)):
# lgb树模型
lgb_reg = lgb.LGBMRegressor(
learning_rate=0.01,
max_depth=-1,
n_estimators=5000,
boosting_type='gbdt',
random_state=2019,
objective='regression',
)
# 切分训练集和预测集
X_train_KFold, X_test_KFold = X_train2[train_index], X_train2[test_index]
y_train_KFold, y_test_KFold = y_train[train_index], y_train[test_index]
# 训练模型
lgb_reg.fit(
X=X_train_KFold,y=y_train_KFold,
eval_set=[(X_train_KFold, y_train_KFold),(X_test_KFold, y_test_KFold)],
eval_names=['Train','Test'],
early_stopping_rounds=100,
eval_metric='MSE',
verbose=50
)
# 训练集预测 测试集预测
y_train_KFold_predict = lgb_reg.predict(X_train_KFold,num_iteration=lgb_reg.best_iteration_)
y_test_KFold_predict = lgb_reg.predict(X_test_KFold,num_iteration=lgb_reg.best_iteration_)
print('第{}折 训练和预测 训练MSE 预测MSE'.format(i))
train_mse = mean_squared_error(y_train_KFold_predict, y_train_KFold)
print('------\n', '训练MSE\n', train_mse, '\n------')
test_mse = mean_squared_error(y_test_KFold_predict, y_test_KFold)
print('------\n', '预测MSE\n', test_mse, '\n------\n')
MSE_DICT['train_mse'].append(train_mse)
MSE_DICT['test_mse'].append(test_mse)
print('------\n', '训练MSE\n', MSE_DICT['train_mse'], '\n', np.mean(MSE_DICT['train_mse']), '\n------')
print('------\n', '预测MSE\n', MSE_DICT['test_mse'], '\n', np.mean(MSE_DICT['test_mse']), '\n------')七、模型融合
导入包
import warnings
warnings.filterwarnings("ignore")
import matplotlib.pyplot as plt
plt.rcParams.update({'figure.max_open_warning': 0})
import seaborn as sns
# modelling
import pandas as pd
import numpy as np
from scipy import stats
from sklearn.model_selection import train_test_split
from sklearn.model_selection import GridSearchCV, RepeatedKFold, cross_val_score,cross_val_predict,KFold
from sklearn.metrics import make_scorer,mean_squared_error
from sklearn.linear_model import LinearRegression, Lasso, Ridge, ElasticNet
from sklearn.svm import LinearSVR, SVR
from sklearn.neighbors import KNeighborsRegressor
from sklearn.ensemble import RandomForestRegressor, GradientBoostingRegressor,AdaBoostRegressor
from xgboost import XGBRegressor
from sklearn.preprocessing import PolynomialFeatures,MinMaxScaler,StandardScaler导入数据
#load_dataset
with open("./zhengqi_train.txt") as fr:
data_train=pd.read_table(fr,sep="\t")
with open("./zhengqi_test.txt") as fr_test:
data_test=pd.read_table(fr_test,sep="\t")合并数据
#merge train_set and test_set
data_train["oringin"]="train"
data_test["oringin"]="test"
data_all=pd.concat([data_train,data_test],axis=0,ignore_index=True)
删除相关特征
data_all.drop(["V5","V9","V11","V17","V22","V28"],axis=1,inplace=True)数据最大最小归一化
# normalise numeric columns
cols_numeric=list(data_all.columns)
cols_numeric.remove("oringin")
def scale_minmax(col):
return (col-col.min())/(col.max()-col.min())
scale_cols = [col for col in cols_numeric if col!='target']
data_all[scale_cols] = data_all[scale_cols].apply(scale_minmax,axis=0)画图:探查特征和标签相关信息
#Check effect of Box-Cox transforms on distributions of continuous variables
fcols = 6
frows = len(cols_numeric)-1
plt.figure(figsize=(4*fcols,4*frows))
i=0
for var in cols_numeric:
if var!='target':
dat = data_all[[var, 'target']].dropna()
i+=1
plt.subplot(frows,fcols,i)
sns.distplot(dat[var] , fit=stats.norm);
plt.title(var+' Original')
plt.xlabel('')
i+=1
plt.subplot(frows,fcols,i)
_=stats.probplot(dat[var], plot=plt)
plt.title('skew='+'{:.4f}'.format(stats.skew(dat[var])))
plt.xlabel('')
plt.ylabel('')
i+=1
plt.subplot(frows,fcols,i)
plt.plot(dat[var], dat['target'],'.',alpha=0.5)
plt.title('corr='+'{:.2f}'.format(np.corrcoef(dat[var], dat['target'])[0][1]))
i+=1
plt.subplot(frows,fcols,i)
trans_var, lambda_var = stats.boxcox(dat[var].dropna()+1)
trans_var = scale_minmax(trans_var)
sns.distplot(trans_var , fit=stats.norm);
plt.title(var+' Tramsformed')
plt.xlabel('')
i+=1
plt.subplot(frows,fcols,i)
_=stats.probplot(trans_var, plot=plt)
plt.title('skew='+'{:.4f}'.format(stats.skew(trans_var)))
plt.xlabel('')
plt.ylabel('')
i+=1
plt.subplot(frows,fcols,i)
plt.plot(trans_var, dat['target'],'.',alpha=0.5)
plt.title('corr='+'{:.2f}'.format(np.corrcoef(trans_var,dat['target'])[0][1]))对特征进行Box-Cox变换,使其满足正态性
cols_transform=data_all.columns[0:-2]
for col in cols_transform:
# transform column
data_all.loc[:,col], _ = stats.boxcox(data_all.loc[:,col]+1)标签数据统计转换后的数据,计算分位数画图展示(基于正态分布)
print(data_all.target.describe())
plt.figure(figsize=(12,4))
plt.subplot(1,2,1)
sns.distplot(data_all.target.dropna() , fit=stats.norm);
plt.subplot(1,2,2)
_=stats.probplot(data_all.target.dropna(), plot=plt)标签数据对数变换数据,使数据更符合正态,画图展示
#Log Transform SalePrice to improve normality
sp = data_train.target
data_train.target1 =np.power(1.5,sp)
print(data_train.target1.describe())
plt.figure(figsize=(12,4))
plt.subplot(1,2,1)
sns.distplot(data_train.target1.dropna(),fit=stats.norm);
plt.subplot(1,2,2)
_=stats.probplot(data_train.target1.dropna(), plot=plt)获取训练和测试数据
# function to get training samples
def get_training_data():
# extract training samples
from sklearn.model_selection import train_test_split
df_train = data_all[data_all["oringin"]=="train"]
df_train["label"]=data_train.target1
# split SalePrice and features
y = df_train.target
X = df_train.drop(["oringin","target","label"],axis=1)
X_train,X_valid,y_train,y_valid=train_test_split(X,y,test_size=0.3,random_state=100)
return X_train,X_valid,y_train,y_valid
# extract test data (without SalePrice)
def get_test_data():
df_test = data_all[data_all["oringin"]=="test"].reset_index(drop=True)
return df_test.drop(["oringin","target"],axis=1)评分函数
from sklearn.metrics import make_scorer
# metric for evaluation
def rmse(y_true, y_pred):
diff = y_pred - y_true
sum_sq = sum(diff**2)
n = len(y_pred)
return np.sqrt(sum_sq/n)
def mse(y_ture,y_pred):
return mean_squared_error(y_ture,y_pred)
# scorer to be used in sklearn model fitting
rmse_scorer = make_scorer(rmse, greater_is_better=False)
mse_scorer = make_scorer(mse, greater_is_better=False)获取异常数据,并画图
# function to detect outliers based on the predictions of a model
def find_outliers(model, X, y, sigma=3):
# predict y values using model
try:
y_pred = pd.Series(model.predict(X), index=y.index)
# if predicting fails, try fitting the model first
except:
model.fit(X,y)
y_pred = pd.Series(model.predict(X), index=y.index)
# calculate residuals between the model prediction and true y values
resid = y - y_pred
mean_resid = resid.mean()
std_resid = resid.std()
# calculate z statistic, define outliers to be where |z|>sigma
z = (resid - mean_resid)/std_resid
outliers = z[abs(z)>sigma].index
# print and plot the results
print('R2=',model.score(X,y))
print('rmse=',rmse(y, y_pred))
print("mse=",mean_squared_error(y,y_pred))
print('---------------------------------------')
print('mean of residuals:',mean_resid)
print('std of residuals:',std_resid)
print('---------------------------------------')
print(len(outliers),'outliers:')
print(outliers.tolist())
plt.figure(figsize=(15,5))
ax_131 = plt.subplot(1,3,1)
plt.plot(y,y_pred,'.')
plt.plot(y.loc[outliers],y_pred.loc[outliers],'ro')
plt.legend(['Accepted','Outlier'])
plt.xlabel('y')
plt.ylabel('y_pred');
ax_132=plt.subplot(1,3,2)
plt.plot(y,y-y_pred,'.')
plt.plot(y.loc[outliers],y.loc[outliers]-y_pred.loc[outliers],'ro')
plt.legend(['Accepted','Outlier'])
plt.xlabel('y')
plt.ylabel('y - y_pred');
ax_133=plt.subplot(1,3,3)
z.plot.hist(bins=50,ax=ax_133)
z.loc[outliers].plot.hist(color='r',bins=50,ax=ax_133)
plt.legend(['Accepted','Outlier'])
plt.xlabel('z')
plt.savefig('outliers.png')
return outliers# get training data
from sklearn.linear_model import Ridge
X_train, X_valid,y_train,y_valid = get_training_data()
test=get_test_data()
# find and remove outliers using a Ridge model
outliers = find_outliers(Ridge(), X_train, y_train)
# permanently remove these outliers from the data
#df_train = data_all[data_all["oringin"]=="train"]
#df_train["label"]=data_train.target1
#df_train=df_train.drop(outliers)
X_outliers=X_train.loc[outliers]
y_outliers=y_train.loc[outliers]
X_t=X_train.drop(outliers)
y_t=y_train.drop(outliers)使用删除异常的数据进行模型训练
def get_trainning_data_omitoutliers():
y1=y_t.copy()
X1=X_t.copy()
return X1,y1采用网格搜索训练模型
from sklearn.preprocessing import StandardScaler
def train_model(model, param_grid=[], X=[], y=[],
splits=5, repeats=5):
# get unmodified training data, unless data to use already specified
if len(y)==0:
X,y = get_trainning_data_omitoutliers()
#poly_trans=PolynomialFeatures(degree=2)
#X=poly_trans.fit_transform(X)
#X=MinMaxScaler().fit_transform(X)
# create cross-validation method
rkfold = RepeatedKFold(n_splits=splits, n_repeats=repeats)
# perform a grid search if param_grid given
if len(param_grid)>0:
# setup grid search parameters
gsearch = GridSearchCV(model, param_grid, cv=rkfold,
scoring="neg_mean_squared_error",
verbose=1, return_train_score=True)
# search the grid
gsearch.fit(X,y)
# extract best model from the grid
model = gsearch.best_estimator_
best_idx = gsearch.best_index_
# get cv-scores for best model
grid_results = pd.DataFrame(gsearch.cv_results_)
cv_mean = abs(grid_results.loc[best_idx,'mean_test_score'])
cv_std = grid_results.loc[best_idx,'std_test_score']
# no grid search, just cross-val score for given model
else:
grid_results = []
cv_results = cross_val_score(model, X, y, scoring="neg_mean_squared_error", cv=rkfold)
cv_mean = abs(np.mean(cv_results))
cv_std = np.std(cv_results)
# combine mean and std cv-score in to a pandas series
cv_score = pd.Series({'mean':cv_mean,'std':cv_std})
# predict y using the fitted model
y_pred = model.predict(X)
# print stats on model performance
print('----------------------')
print(model)
print('----------------------')
print('score=',model.score(X,y))
print('rmse=',rmse(y, y_pred))
print('mse=',mse(y, y_pred))
print('cross_val: mean=',cv_mean,', std=',cv_std)
# residual plots
y_pred = pd.Series(y_pred,index=y.index)
resid = y - y_pred
mean_resid = resid.mean()
std_resid = resid.std()
z = (resid - mean_resid)/std_resid
n_outliers = sum(abs(z)>3)
plt.figure(figsize=(15,5))
ax_131 = plt.subplot(1,3,1)
plt.plot(y,y_pred,'.')
plt.xlabel('y')
plt.ylabel('y_pred');
plt.title('corr = {:.3f}'.format(np.corrcoef(y,y_pred)[0][1]))
ax_132=plt.subplot(1,3,2)
plt.plot(y,y-y_pred,'.')
plt.xlabel('y')
plt.ylabel('y - y_pred');
plt.title('std resid = {:.3f}'.format(std_resid))
ax_133=plt.subplot(1,3,3)
z.plot.hist(bins=50,ax=ax_133)
plt.xlabel('z')
plt.title('{:.0f} samples with z>3'.format(n_outliers))
return model, cv_score, grid_results# places to store optimal models and scores
opt_models = dict()
score_models = pd.DataFrame(columns=['mean','std'])
# no. k-fold splits
splits=5
# no. k-fold iterations
repeats=5岭回归
model = 'Ridge'
opt_models[model] = Ridge()
alph_range = np.arange(0.25,6,0.25)
param_grid = {'alpha': alph_range}
opt_models[model],cv_score,grid_results = train_model(opt_models[model], param_grid=param_grid,
splits=splits, repeats=repeats)
cv_score.name = model
score_models = score_models.append(cv_score)
plt.figure()
plt.errorbar(alph_range, abs(grid_results['mean_test_score']),
abs(grid_results['std_test_score'])/np.sqrt(splits*repeats))
plt.xlabel('alpha')
plt.ylabel('score')Lasso回归
model = 'Lasso'
opt_models[model] = Lasso()
alph_range = np.arange(1e-4,1e-3,4e-5)
param_grid = {'alpha': alph_range}
opt_models[model], cv_score, grid_results = train_model(opt_models[model], param_grid=param_grid,
splits=splits, repeats=repeats)
cv_score.name = model
score_models = score_models.append(cv_score)
plt.figure()
plt.errorbar(alph_range, abs(grid_results['mean_test_score']),abs(grid_results['std_test_score'])/np.sqrt(splits*repeats))
plt.xlabel('alpha')
plt.ylabel('score')ElasticNet回归
model ='ElasticNet'
opt_models[model] = ElasticNet()
param_grid = {'alpha': np.arange(1e-4,1e-3,1e-4),
'l1_ratio': np.arange(0.1,1.0,0.1),
'max_iter':[100000]}
opt_models[model], cv_score, grid_results = train_model(opt_models[model], param_grid=param_grid,
splits=splits, repeats=1)
cv_score.name = model
score_models = score_models.append(cv_score)SVR回归
model='LinearSVR'
opt_models[model] = LinearSVR()
crange = np.arange(0.1,1.0,0.1)
param_grid = {'C':crange,
'max_iter':[1000]}
opt_models[model], cv_score, grid_results = train_model(opt_models[model], param_grid=param_grid,
splits=splits, repeats=repeats)
cv_score.name = model
score_models = score_models.append(cv_score)
plt.figure()
plt.errorbar(crange, abs(grid_results['mean_test_score']),abs(grid_results['std_test_score'])/np.sqrt(splits*repeats))
plt.xlabel('C')
plt.ylabel('score')K最近邻
model = 'KNeighbors'
opt_models[model] = KNeighborsRegressor()
param_grid = {'n_neighbors':np.arange(3,11,1)}
opt_models[model], cv_score, grid_results = train_model(opt_models[model], param_grid=param_grid,
splits=splits, repeats=1)
cv_score.name = model
score_models = score_models.append(cv_score)
plt.figure()
plt.errorbar(np.arange(3,11,1), abs(grid_results['mean_test_score']),abs(grid_results['std_test_score'])/np.sqrt(splits*1))
plt.xlabel('n_neighbors')
plt.ylabel('score')GBDT模型
model = 'GradientBoosting'
opt_models[model] = GradientBoostingRegressor()
param_grid = {'n_estimators':[150,250,350],
'max_depth':[1,2,3],
'min_samples_split':[5,6,7]}
opt_models[model], cv_score, grid_results = train_model(opt_models[model], param_grid=param_grid,
splits=splits, repeats=1)
cv_score.name = model
score_models = score_models.append(cv_score)XGB模型
model = 'XGB'
opt_models[model] = XGBRegressor()
param_grid = {'n_estimators':[100,200,300,400,500],
'max_depth':[1,2,3],
}
opt_models[model], cv_score,grid_results = train_model(opt_models[model], param_grid=param_grid,
splits=splits, repeats=1)
cv_score.name = model
score_models = score_models.append(cv_score)随机森林模型
model = 'RandomForest'
opt_models[model] = RandomForestRegressor()
param_grid = {'n_estimators':[100,150,200],
'max_features':[8,12,16,20,24],
'min_samples_split':[2,4,6]}
opt_models[model], cv_score, grid_results = train_model(opt_models[model], param_grid=param_grid,
splits=5, repeats=1)
cv_score.name = model
score_models = score_models.append(cv_score)模型预测--多模型Begging
def model_predict(test_data,test_y=[],stack=False):
#poly_trans=PolynomialFeatures(degree=2)
#test_data1=poly_trans.fit_transform(test_data)
#test_data=MinMaxScaler().fit_transform(test_data)
i=0
y_predict_total=np.zeros((test_data.shape[0],))
for model in opt_models.keys():
if model!="LinearSVR" and model!="KNeighbors":
y_predict=opt_models[model].predict(test_data)
y_predict_total+=y_predict
i+=1
if len(test_y)>0:
print("{}_mse:".format(model),mean_squared_error(y_predict,test_y))
y_predict_mean=np.round(y_predict_total/i,3)
if len(test_y)>0:
print("mean_mse:",mean_squared_error(y_predict_mean,test_y))
else:
y_predict_mean=pd.Series(y_predict_mean)
return y_predict_meanBagging预测
model_predict(X_valid,y_valid)模型融合Stacking
模型融合stacking简单示例
import numpy as np
import matplotlib.pyplot as plt
import matplotlib.gridspec as gridspec
import itertools
from sklearn.linear_model import LogisticRegression
from sklearn.svm import SVC
from sklearn.ensemble import RandomForestClassifier
##主要使用pip install mlxtend安装mlxtend
from mlxtend.classifier import EnsembleVoteClassifier
from mlxtend.data import iris_data
from mlxtend.plotting import plot_decision_regions
%matplotlib inline
# Initializing Classifiers
clf1 = LogisticRegression(random_state=0)
clf2 = RandomForestClassifier(random_state=0)
clf3 = SVC(random_state=0, probability=True)
eclf = EnsembleVoteClassifier(clfs=[clf1, clf2, clf3], weights=[2, 1, 1], voting='soft')
# Loading some example data
X, y = iris_data()
X = X[:,[0, 2]]
# Plotting Decision Regions
gs = gridspec.GridSpec(2, 2)
fig = plt.figure(figsize=(10, 8))
for clf, lab, grd in zip([clf1, clf2, clf3, eclf],
['Logistic Regression', 'Random Forest', 'RBF kernel SVM', 'Ensemble'],
itertools.product([0, 1], repeat=2)):
clf.fit(X, y)
ax = plt.subplot(gs[grd[0], grd[1]])
fig = plot_decision_regions(X=X, y=y, clf=clf, legend=2)
plt.title(lab)
plt.show()工业蒸汽多模型融合stacking
from sklearn.model_selection import train_test_split
import pandas as pd
import numpy as np
from scipy import sparse
import xgboost
import lightgbm
from sklearn.ensemble import RandomForestRegressor,AdaBoostRegressor,GradientBoostingRegressor,ExtraTreesRegressor
from sklearn.linear_model import LinearRegression
from sklearn.metrics import mean_squared_error
def stacking_reg(clf,train_x,train_y,test_x,clf_name,kf,label_split=None):
train=np.zeros((train_x.shape[0],1))
test=np.zeros((test_x.shape[0],1))
test_pre=np.empty((folds,test_x.shape[0],1))
cv_scores=[]
for i,(train_index,test_index) in enumerate(kf.split(train_x,label_split)):
tr_x=train_x[train_index]
tr_y=train_y[train_index]
te_x=train_x[test_index]
te_y = train_y[test_index]
if clf_name in ["rf","ada","gb","et","lr","lsvc","knn"]:
clf.fit(tr_x,tr_y)
pre=clf.predict(te_x).reshape(-1,1)
train[test_index]=pre
test_pre[i,:]=clf.predict(test_x).reshape(-1,1)
cv_scores.append(mean_squared_error(te_y, pre))
elif clf_name in ["xgb"]:
train_matrix = clf.DMatrix(tr_x, label=tr_y, missing=-1)
test_matrix = clf.DMatrix(te_x, label=te_y, missing=-1)
z = clf.DMatrix(test_x, label=te_y, missing=-1)
params = {'booster': 'gbtree',
'eval_metric': 'rmse',
'gamma': 1,
'min_child_weight': 1.5,
'max_depth': 5,
'lambda': 10,
'subsample': 0.7,
'colsample_bytree': 0.7,
'colsample_bylevel': 0.7,
'eta': 0.03,
'tree_method': 'exact',
'seed': 2017,
'nthread': 12
}
num_round = 10000
early_stopping_rounds = 100
watchlist = [(train_matrix, 'train'),
(test_matrix, 'eval')
]
if test_matrix:
model = clf.train(params, train_matrix, num_boost_round=num_round,evals=watchlist,
early_stopping_rounds=early_stopping_rounds
)
pre= model.predict(test_matrix,ntree_limit=model.best_ntree_limit).reshape(-1,1)
train[test_index]=pre
test_pre[i, :]= model.predict(z, ntree_limit=model.best_ntree_limit).reshape(-1,1)
cv_scores.append(mean_squared_error(te_y, pre))
elif clf_name in ["lgb"]:
train_matrix = clf.Dataset(tr_x, label=tr_y)
test_matrix = clf.Dataset(te_x, label=te_y)
#z = clf.Dataset(test_x, label=te_y)
#z=test_x
params = {
'boosting_type': 'gbdt',
'objective': 'regression_l2',
'metric': 'mse',
'min_child_weight': 1.5,
'num_leaves': 2**5,
'lambda_l2': 10,
'subsample': 0.7,
'colsample_bytree': 0.7,
'colsample_bylevel': 0.7,
'learning_rate': 0.03,
'tree_method': 'exact',
'seed': 2017,
'nthread': 12,
'silent': True,
}
num_round = 10000
early_stopping_rounds = 100
if test_matrix:
model = clf.train(params, train_matrix,num_round,valid_sets=test_matrix,
early_stopping_rounds=early_stopping_rounds
)
pre= model.predict(te_x,num_iteration=model.best_iteration).reshape(-1,1)
train[test_index]=pre
test_pre[i, :]= model.predict(test_x, num_iteration=model.best_iteration).reshape(-1,1)
cv_scores.append(mean_squared_error(te_y, pre))
else:
raise IOError("Please add new clf.")
print("%s now score is:"%clf_name,cv_scores)
test[:]=test_pre.mean(axis=0)
print("%s_score_list:"%clf_name,cv_scores)
print("%s_score_mean:"%clf_name,np.mean(cv_scores))
return train.reshape(-1,1),test.reshape(-1,1)模型融合stacking基学习器
def rf_reg(x_train, y_train, x_valid, kf, label_split=None):
randomforest = RandomForestRegressor(n_estimators=600, max_depth=20, n_jobs=-1, random_state=2017, max_features="auto",verbose=1)
rf_train, rf_test = stacking_reg(randomforest, x_train, y_train, x_valid, "rf", kf, label_split=label_split)
return rf_train, rf_test,"rf_reg"
def ada_reg(x_train, y_train, x_valid, kf, label_split=None):
adaboost = AdaBoostRegressor(n_estimators=30, random_state=2017, learning_rate=0.01)
ada_train, ada_test = stacking_reg(adaboost, x_train, y_train, x_valid, "ada", kf, label_split=label_split)
return ada_train, ada_test,"ada_reg"
def gb_reg(x_train, y_train, x_valid, kf, label_split=None):
gbdt = GradientBoostingRegressor(learning_rate=0.04, n_estimators=100, subsample=0.8, random_state=2017,max_depth=5,verbose=1)
gbdt_train, gbdt_test = stacking_reg(gbdt, x_train, y_train, x_valid, "gb", kf, label_split=label_split)
return gbdt_train, gbdt_test,"gb_reg"
def et_reg(x_train, y_train, x_valid, kf, label_split=None):
extratree = ExtraTreesRegressor(n_estimators=600, max_depth=35, max_features="auto", n_jobs=-1, random_state=2017,verbose=1)
et_train, et_test = stacking_reg(extratree, x_train, y_train, x_valid, "et", kf, label_split=label_split)
return et_train, et_test,"et_reg"
def lr_reg(x_train, y_train, x_valid, kf, label_split=None):
lr_reg=LinearRegression(n_jobs=-1)
lr_train, lr_test = stacking_reg(lr_reg, x_train, y_train, x_valid, "lr", kf, label_split=label_split)
return lr_train, lr_test, "lr_reg"
def xgb_reg(x_train, y_train, x_valid, kf, label_split=None):
xgb_train, xgb_test = stacking_reg(xgboost, x_train, y_train, x_valid, "xgb", kf, label_split=label_split)
return xgb_train, xgb_test,"xgb_reg"
def lgb_reg(x_train, y_train, x_valid, kf, label_split=None):
lgb_train, lgb_test = stacking_reg(lightgbm, x_train, y_train, x_valid, "lgb", kf, label_split=label_split)
return lgb_train, lgb_test,"lgb_reg"模型融合stacking预测
def stacking_pred(x_train, y_train, x_valid, kf, clf_list, label_split=None, clf_fin="lgb", if_concat_origin=True):
for k, clf_list in enumerate(clf_list):
clf_list = [clf_list]
column_list = []
train_data_list=[]
test_data_list=[]
for clf in clf_list:
train_data,test_data,clf_name=clf(x_train, y_train, x_valid, kf, label_split=label_split)
train_data_list.append(train_data)
test_data_list.append(test_data)
column_list.append("clf_%s" % (clf_name))
train = np.concatenate(train_data_list, axis=1)
test = np.concatenate(test_data_list, axis=1)
if if_concat_origin:
train = np.concatenate([x_train, train], axis=1)
test = np.concatenate([x_valid, test], axis=1)
print(x_train.shape)
print(train.shape)
print(clf_name)
print(clf_name in ["lgb"])
if clf_fin in ["rf","ada","gb","et","lr","lsvc","knn"]:
if clf_fin in ["rf"]:
clf = RandomForestRegressor(n_estimators=600, max_depth=20, n_jobs=-1, random_state=2017, max_features="auto",verbose=1)
elif clf_fin in ["ada"]:
clf = AdaBoostRegressor(n_estimators=30, random_state=2017, learning_rate=0.01)
elif clf_fin in ["gb"]:
clf = GradientBoostingRegressor(learning_rate=0.04, n_estimators=100, subsample=0.8, random_state=2017,max_depth=5,verbose=1)
elif clf_fin in ["et"]:
clf = ExtraTreesRegressor(n_estimators=600, max_depth=35, max_features="auto", n_jobs=-1, random_state=2017,verbose=1)
elif clf_fin in ["lr"]:
clf = LinearRegression(n_jobs=-1)
clf.fit(train, y_train)
pre = clf.predict(test).reshape(-1,1)
return pred
elif clf_fin in ["xgb"]:
clf = xgboost
train_matrix = clf.DMatrix(train, label=y_train, missing=-1)
test_matrix = clf.DMatrix(train, label=y_train, missing=-1)
params = {'booster': 'gbtree',
'eval_metric': 'rmse',
'gamma': 1,
'min_child_weight': 1.5,
'max_depth': 5,
'lambda': 10,
'subsample': 0.7,
'colsample_bytree': 0.7,
'colsample_bylevel': 0.7,
'eta': 0.03,
'tree_method': 'exact',
'seed': 2017,
'nthread': 12
}
num_round = 10000
early_stopping_rounds = 100
watchlist = [(train_matrix, 'train'),
(test_matrix, 'eval')
]
model = clf.train(params, train_matrix, num_boost_round=num_round,evals=watchlist,
early_stopping_rounds=early_stopping_rounds
)
pre = model.predict(test,ntree_limit=model.best_ntree_limit).reshape(-1,1)
return pre
elif clf_fin in ["lgb"]:
print(clf_name)
clf = lightgbm
train_matrix = clf.Dataset(train, label=y_train)
test_matrix = clf.Dataset(train, label=y_train)
params = {
'boosting_type': 'gbdt',
'objective': 'regression_l2',
'metric': 'mse',
'min_child_weight': 1.5,
'num_leaves': 2**5,
'lambda_l2': 10,
'subsample': 0.7,
'colsample_bytree': 0.7,
'colsample_bylevel': 0.7,
'learning_rate': 0.03,
'tree_method': 'exact',
'seed': 2017,
'nthread': 12,
'silent': True,
}
num_round = 10000
early_stopping_rounds = 100
model = clf.train(params, train_matrix,num_round,valid_sets=test_matrix,
early_stopping_rounds=early_stopping_rounds
)
print('pred')
pre = model.predict(test,num_iteration=model.best_iteration).reshape(-1,1)
print(pre)
return pre加载数据
# #load_dataset
with open("./zhengqi_train.txt") as fr:
data_train=pd.read_table(fr,sep="\t")
with open("./zhengqi_test.txt") as fr_test:
data_test=pd.read_table(fr_test,sep="\t")k折交叉验证
from sklearn.model_selection import StratifiedKFold, KFold
folds = 5
seed = 1
kf = KFold(n_splits=5, shuffle=True, random_state=0)训练集和测试集数据
x_train = data_train[data_test.columns].values
x_valid = data_test[data_test.columns].values
y_train = data_train['target'].values使用lr_reg和lgb_reg进行融合预测
clf_list = [lr_reg, lgb_reg]
#clf_list = [lr_reg, rf_reg]
##很容易过拟合
pred = stacking_pred(x_train, y_train, x_valid, kf, clf_list, label_split=None, clf_fin="lgb", if_concat_origin=True)总结
以上是有关工业蒸汽预测的案例。





















 1090
1090











 被折叠的 条评论
为什么被折叠?
被折叠的 条评论
为什么被折叠?








