最全的Linux教程,Linux从入门到精通
======================
-
linux从入门到精通(第2版)
-
Linux系统移植
-
Linux驱动开发入门与实战
-
LINUX 系统移植 第2版
-
Linux开源网络全栈详解 从DPDK到OpenFlow

第一份《Linux从入门到精通》466页
====================
内容简介
====
本书是获得了很多读者好评的Linux经典畅销书**《Linux从入门到精通》的第2版**。本书第1版出版后曾经多次印刷,并被51CTO读书频道评为“最受读者喜爱的原创IT技术图书奖”。本书第﹖版以最新的Ubuntu 12.04为版本,循序渐进地向读者介绍了Linux 的基础应用、系统管理、网络应用、娱乐和办公、程序开发、服务器配置、系统安全等。本书附带1张光盘,内容为本书配套多媒体教学视频。另外,本书还为读者提供了大量的Linux学习资料和Ubuntu安装镜像文件,供读者免费下载。

本书适合广大Linux初中级用户、开源软件爱好者和大专院校的学生阅读,同时也非常适合准备从事Linux平台开发的各类人员。
需要《Linux入门到精通》、《linux系统移植》、《Linux驱动开发入门实战》、《Linux开源网络全栈》电子书籍及教程的工程师朋友们劳烦您转发+评论
网上学习资料一大堆,但如果学到的知识不成体系,遇到问题时只是浅尝辄止,不再深入研究,那么很难做到真正的技术提升。
一个人可以走的很快,但一群人才能走的更远!不论你是正从事IT行业的老鸟或是对IT行业感兴趣的新人,都欢迎加入我们的的圈子(技术交流、学习资源、职场吐槽、大厂内推、面试辅导),让我们一起学习成长!
&& wget http://bcb.unl.edu/dbCAN2/download/Databases/V11/CAZyDB.08062022.fa && diamond makedb --in CAZyDB.08062022.fa -d CAZy \
&& wget https://bcb.unl.edu/dbCAN2/download/Databases/V11/dbCAN-HMMdb-V11.txt && mv dbCAN-HMMdb-V11.txt dbCAN.txt && hmmpress dbCAN.txt \
&& wget https://bcb.unl.edu/dbCAN2/download/Databases/V11/tcdb.fa && diamond makedb --in tcdb.fa -d tcdb \
&& wget http://bcb.unl.edu/dbCAN2/download/Databases/V11/tf-1.hmm && hmmpress tf-1.hmm \
&& wget http://bcb.unl.edu/dbCAN2/download/Databases/V11/tf-2.hmm && hmmpress tf-2.hmm \
&& wget https://bcb.unl.edu/dbCAN2/download/Databases/V11/stp.hmm && hmmpress stp.hmm \
&& cd ../ && wget http://bcb.unl.edu/dbCAN2/download/Samples/EscheriaColiK12MG1655.fna \
&& wget http://bcb.unl.edu/dbCAN2/download/Samples/EscheriaColiK12MG1655.faa \
&& wget http://bcb.unl.edu/dbCAN2/download/Samples/EscheriaColiK12MG1655.gff
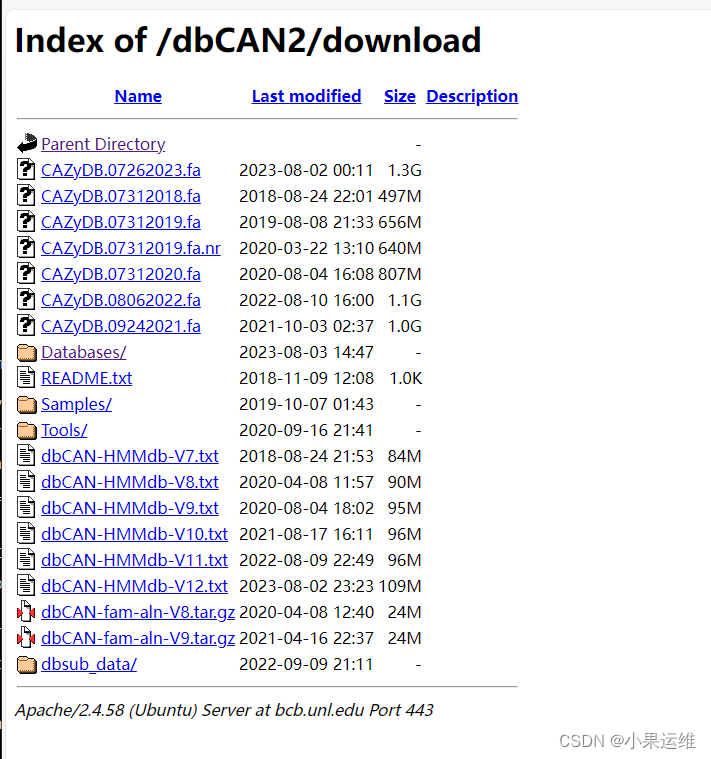
手动下载位置:[Index of /dbCAN2/download (unl.edu)]( )")
SignalP数据库下载和配置
文章:[Predicting Secretory Proteins with SignalP | SpringerLink]( )
[SignalP 4.1 - DTU Health Tech - Bioinformatic Services]( )
**需要填写邮箱信息同意后才会发送限时链接(4小时内有效)到对应邮箱**
**当然大家可以直接在网上丢个fasta文件,选择参数后提交在线的注释任务。**
## 3、使用run\_dbcan
### 帮助信息:
Required arguments:
inputFile User input file. Must be in FASTA format.
{protein,prok,meta} Type of sequence input. protein=proteome; prok=prokaryote; meta=metagenome
optional arguments:
-h, --help show this help message and exit
–dbCANFile DBCANFILE
Indicate the file name of HMM database such as dbCAN.txt, please use the newest one from dbCAN2 website.
–dia_eval DIA_EVAL DIAMOND E Value
–dia_cpu DIA_CPU Number of CPU cores that DIAMOND is allowed to use
–hmm_eval HMM_EVAL HMMER E Value
–hmm_cov HMM_COV HMMER Coverage val
–hmm_cpu HMM_CPU Number of CPU cores that HMMER is allowed to use
–out_pre OUT_PRE Output files prefix
–out_dir OUT_DIR Output directory
–db_dir DB_DIR Database directory
–tools {hmmer,diamond,dbcansub,all} [{hmmer,diamond,dbcansub,all} …], -t {hmmer,diamond,dbcansub,all} [{hmmer,diamond,dbcansub,all} …]
Choose a combination of tools to run
–use_signalP USE_SIGNALP
Use signalP or not, remember, you need to setup signalP tool first. Because of signalP license, Docker version does not have signalP.
–signalP_path SIGNALP_PATH, -sp SIGNALP_PATH
The path for signalp. Default location is signalp
–gram {p,n,all}, -g {p,n,all}
Choose gram+§ or gram-(n) for proteome/prokaryote nucleotide, which are params of SingalP, only if user use singalP
-v VERSION, --version VERSION
dbCAN-sub parameters:
–dbcan_thread DBCAN_THREAD, -dt DBCAN_THREAD
–tf_eval TF_EVAL tf.hmm HMMER E Value
–tf_cov TF_COV tf.hmm HMMER Coverage val
–tf_cpu TF_CPU tf.hmm Number of CPU cores that HMMER is allowed to use
–stp_eval STP_EVAL stp.hmm HMMER E Value
–stp_cov STP_COV stp.hmm HMMER Coverage val
–stp_cpu STP_CPU stp.hmm Number of CPU cores that HMMER is allowed to use
CGC_Finder parameters:
–cluster CLUSTER, -c CLUSTER
Predict CGCs via CGCFinder. This argument requires an auxillary locations file if a protein input is being used
–cgc_dis CGC_DIS CGCFinder Distance value
–cgc_sig_genes {tf,tp,stp,tp+tf,tp+stp,tf+stp,all}
CGCFinder Signature Genes value
CGC_Substrate parameters:
–cgc_substrate run cgc substrate prediction?
–pul PUL dbCAN-PUL PUL.faa
-o OUT, --out OUT
-w WORKDIR, --workdir WORKDIR
-env ENV, --env ENV
-oecami, --oecami out eCAMI prediction intermediate result?
-odbcanpul, --odbcanpul
output dbCAN-PUL prediction intermediate result?
dbCAN-PUL homologous searching parameters:
how to define homologous gene hits and PUL hits
-upghn UNIQ_PUL_GENE_HIT_NUM, --uniq_pul_gene_hit_num UNIQ_PUL_GENE_HIT_NUM
-uqcgn UNIQ_QUERY_CGC_GENE_NUM, --uniq_query_cgc_gene_num UNIQ_QUERY_CGC_GENE_NUM
-cpn CAZYME_PAIR_NUM, --CAZyme_pair_num CAZYME_PAIR_NUM
-tpn TOTAL_PAIR_NUM, --total_pair_num TOTAL_PAIR_NUM
-ept EXTRA_PAIR_TYPE, --extra_pair_type EXTRA_PAIR_TYPE
None[TC-TC,STP-STP]. Some like sigunature hits
-eptn EXTRA_PAIR_TYPE_NUM, --extra_pair_type_num EXTRA_PAIR_TYPE_NUM
specify signature pair cutoff.1,2
-iden IDENTITY_CUTOFF, --identity_cutoff IDENTITY_CUTOFF
identity to identify a homologous hit
-cov COVERAGE_CUTOFF, --coverage_cutoff COVERAGE_CUTOFF
query coverage cutoff to identify a homologous hit
-bsc BITSCORE_CUTOFF, --bitscore_cutoff BITSCORE_CUTOFF
bitscore cutoff to identify a homologous hit
-evalue EVALUE_CUTOFF, --evalue_cutoff EVALUE_CUTOFF
evalue cutoff to identify a homologous hit
dbCAN-sub major voting parameters:
how to define dbsub hits and dbCAN-sub subfamily substrate
-hmmcov HMMCOV, --hmmcov HMMCOV
-hmmevalue HMMEVALUE, --hmmevalue HMMEVALUE
-ndsc NUM_OF_DOMAINS_SUBSTRATE_CUTOFF, --num_of_domains_substrate_cutoff NUM_OF_DOMAINS_SUBSTRATE_CUTOFF
define how many domains share substrates in a CGC, one protein may include several subfamily domains.
-npsc NUM_OF_PROTEIN_SUBSTRATE_CUTOFF, --num_of_protein_substrate_cutoff NUM_OF_PROTEIN_SUBSTRATE_CUTOFF
define how many sequences share substrates in a CGC, one protein may include several subfamily domains.
-subs SUBSTRATE_SCORS, --substrate_scors SUBSTRATE_SCORS
each cgc contains with substrate must more than this value
### 命令及结果参考
#参考格式
run_dbcan [inputFile] [inputType] [-c AuxillaryFile] [-t Tools]
#结果说明
uniInput - The unified input file for the rest of the tools
(created by prodigal if a nucleotide sequence was used)
dbsub.out - the output from the dbCAN_sub run
diamond.out - the output from the diamond blast
hmmer.out - the output from the hmmer run
tf.out - the output from the diamond blast predicting TF’s for CGCFinder
tc.out - the output from the diamond blast predicting TC’s for CGCFinder
cgc.gff - GFF input file for CGCFinder
cgc.out - ouput from the CGCFinder run
overview.txt - Details the CAZyme predictions across the three tools with signalp results
###说的都很清楚了,就不重复了,英文可以chatgpt或者百度吧
示例:
run_dbcan EscheriaColiK12MG1655.fna prok --out_dir output_EscheriaColiK12MG1655
run_dbcan EscheriaColiK12MG1655.faa protein --out_dir output_EscheriaColiK12MG1655
run_dbcan EscheriaColiK12MG1655.fna prok -c cluster --out_dir output_EscheriaColiK12MG1655
run_dbcan EscheriaColiK12MG1655.faa protein -c EscheriaColiK12MG1655.gff --out_dir output_EscheriaColiK12MG1655
## 手动注释CAZyDB
### 1、下载指定文件的数据库文件,注意下载最新版本:
###中间07312020表示2020年7月31日的版本,大家可以浏览download目录查看确认最新版
wget -c http://bcb.unl.edu/dbCAN2/download/CAZyDB.07312020.fa
wget -c http://bcb.unl.edu/dbCAN2/download/Databases/CAZyDB.07302020.fam-activities.txt
### 2、使用diamond工具进行快速比对
#基于fasta文件生成diamond比对参考数据库
diamond makedb --in CAZyDB.07312020.fa --db CAZyDB.07312020
提取fam对应注释
grep -v ‘#’ CAZyDB.07302020.fam-activities.txt |sed ‘s/ //’| sed ‘1 i CAZy\tDescription’ > CAZy_description.txt
###位置 /database/CAZyDB
diamond blastp --db /database/CAZyDB/CAZyDB.07312020 --query out_pro.fa --threads 10 -e 1e-5 --outfmt 6 --max-target-seqs 1 --quiet --out ./gene_diamond.f6
提取基因与dbcan分类对应表
perl ./format_dbcan2list.pl -i gene_diamond.f6 -o gene.list
#按对应表累计丰度
python ./summarizeAbundance.py -i gene.count -m gene.list -c 2 -s ‘,’ -n raw -o ./TPM
这里面format\_dbcan2list.pl和summarizeAbundance.py的来源是来自刘永鑫文章和github代码仓库,后面有时间再给大家做详细介绍,或者大家看相关文章自己研究:
[https://doi.org/10.1002/imt2.83]( )
**网上学习资料一大堆,但如果学到的知识不成体系,遇到问题时只是浅尝辄止,不再深入研究,那么很难做到真正的技术提升。**
**[需要这份系统化的资料的朋友,可以点击这里获取!](https://bbs.csdn.net/topics/618635766)**
**一个人可以走的很快,但一群人才能走的更远!不论你是正从事IT行业的老鸟或是对IT行业感兴趣的新人,都欢迎加入我们的的圈子(技术交流、学习资源、职场吐槽、大厂内推、面试辅导),让我们一起学习成长!**






















 2457
2457











 被折叠的 条评论
为什么被折叠?
被折叠的 条评论
为什么被折叠?








