1写在前面
今天发现MendelianRandomization包更新v0.9了。😜
其实也算不上更新。🫠
跟大家一起分享一下这个包做MR的用法吧。🤩
还有一个包就是TwoSampleMR,大家有兴趣可以去学一下。😅
2用到的包
rm(list = ls())
# install.packages("MendelianRandomization")
library(MendelianRandomization)
library(tidyverse)
3示例数据
这里需要说明一下哦,不是所有的参数都需要填的,这个需要根据你后面用到的方法。😘
比如说一些方法不需要指定bxse。😜
mr_ivw函数是将BXSE设置为零的情况下运行的。😀
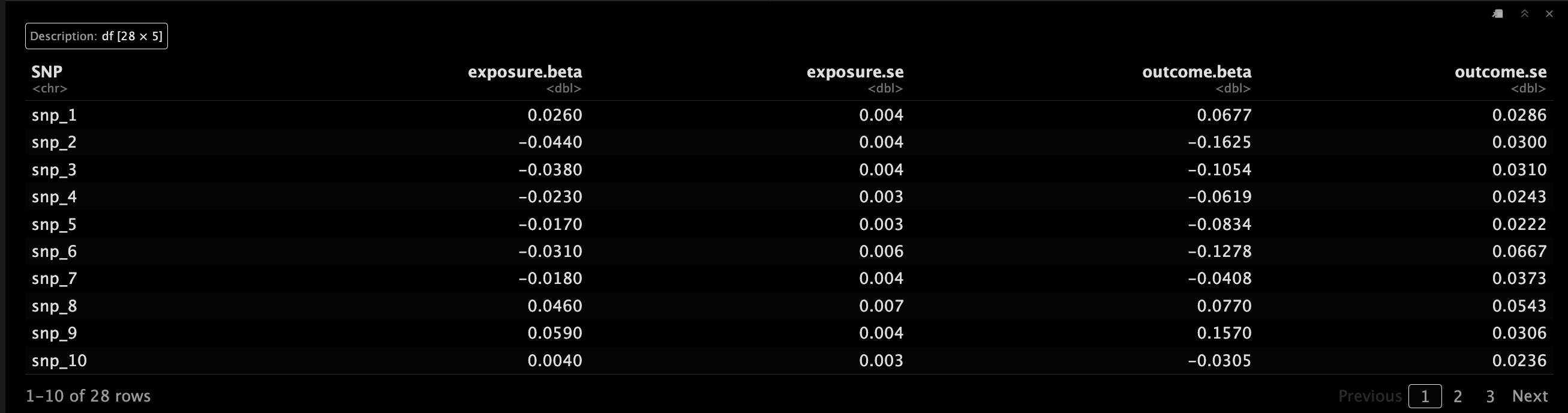
示例一
betaX是来自每个SNP的暴露因素的回归分析的β系数,betaXse是标准误。😘
betaY是每个SNP的结局时间的回归分析的β系数数,betaYse是标准误。🥳
MRInputObject <- mr_input(bx = ldlc,
bxse = ldlcse,
by = chdlodds,
byse = chdloddsse)
MRInputObject
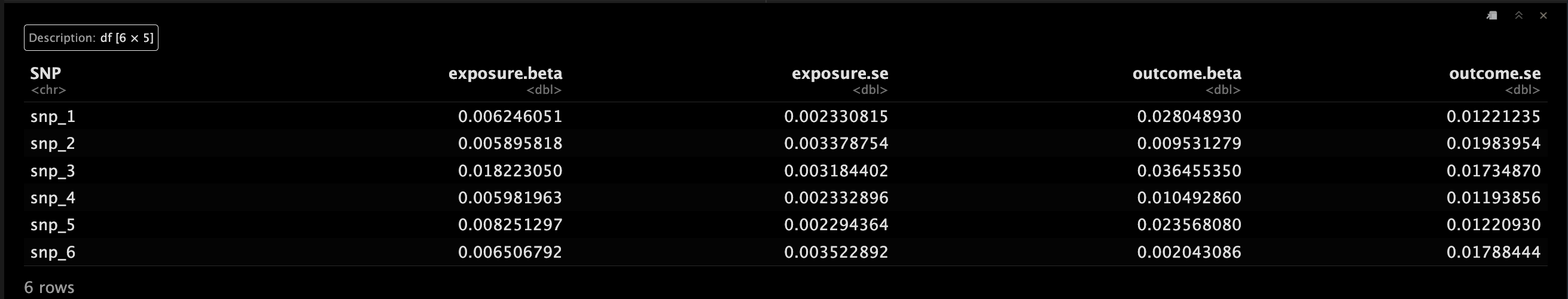
示例二
如果SNP是存在存在相关性,这个时候可以通过corr = 加入哦。😯
MRInputObject.cor <- mr_input(bx = calcium,
bxse = calciumse,
by = fastgluc,
byse = fastglucse,
corr = calc.rho)
MRInputObject.cor
这里的两个data是出自下面2个原文,大家可以去读一下,试试复现:👇
1️⃣ Burgess S, Scott RA, Timpson NJ, Davey Smith G, Thompson SG; EPIC- InterAct Consortium. Using published data in Mendelian randomization: a blueprint for efficient identification of causal risk factors. Eur J Epidemiol. 2015 Jul;30(7):543-52. doi: 10.1007/s10654-015-0011-zIF:
13.6 Q1 . Epub 2015 Mar 15. PMID: 25773750
IF: 13.6 Q1 ; PMCID: PMC4516908
IF: 13.6 Q1 .
2️⃣ Waterworth DM, Ricketts SL, Song K, Chen L, Zhao JH, Ripatti S, Aulchenko YS, Zhang W, Yuan X, Lim N, Luan J, Ashford S, Wheeler E, Young EH, Hadley D, Thompson JR, Braund PS, Johnson T, Struchalin M, Surakka I, Luben R, Khaw KT, Rodwell SA, Loos RJ, Boekholdt SM, Inouye M, Deloukas P, Elliott P, Schlessinger D, Sanna S, Scuteri A, Jackson A, Mohlke KL, Tuomilehto J, Roberts R, Stewart A, Kesäniemi YA, Mahley RW, Grundy SM; Wellcome Trust Case Control Consortium; McArdle W, Cardon L, Waeber G, Vollenweider P, Chambers JC, Boehnke M, Abecasis GR, Salomaa V, Järvelin MR, Ruokonen A, Barroso I, Epstein SE, Hakonarson HH, Rader DJ, Reilly MP, Witteman JC, Hall AS, Samani NJ, Strachan DP, Barter P, van Duijn CM, Kooner JS, Peltonen L, Wareham NJ, McPherson R, Mooser V, Sandhu MS. Genetic variants influencing circulating lipid levels and risk of coronary artery disease. Arterioscler Thromb Vasc Biol. 2010 Nov;30(11):2264-76. doi: 10.1161/ATVBAHA.109.201020
IF: 8.7 Q1 . Epub 2010 Sep 23. PMID: 20864672
IF: 8.7 Q1 ; PMCID: PMC3891568
IF: 8.7 Q1 .
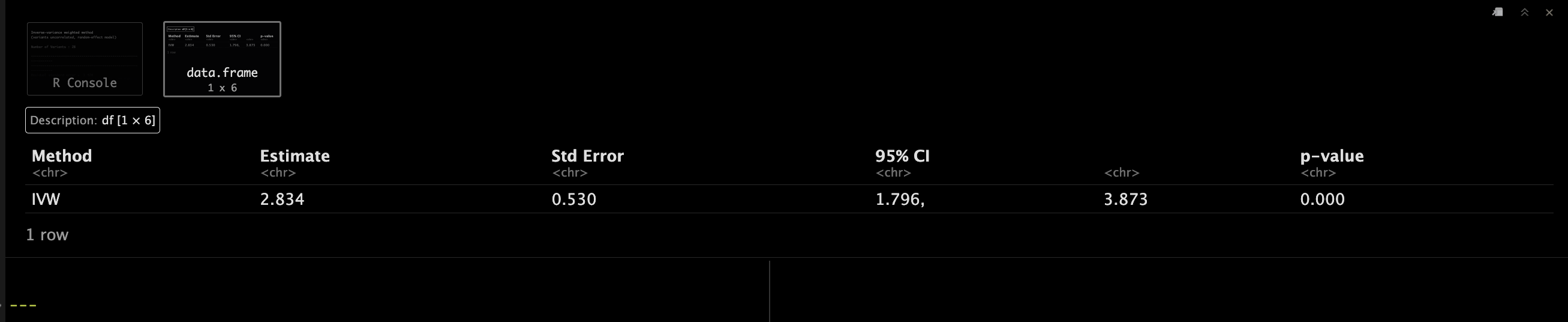
4方法一 Inverse-variance weighted method
在MR的因果推断中,我们常用四种方法,inverse-variance weighted method, median-based method,MR-Egger method,还有Maximum likelihood method。😘
我们注意逐一介绍下。🔽
IVWObject <- mr_ivw(MRInputObject,
model = "default",
robust = FALSE,
penalized = FALSE,
correl = FALSE,
weights = "simple",
psi = 0,
distribution = "normal",
alpha = 0.05)
IVWObject <- mr_ivw(mr_input(bx = ldlc,
bxse = ldlcse,
by = chdlodds,
byse = chdloddsse)
)
IVWObject
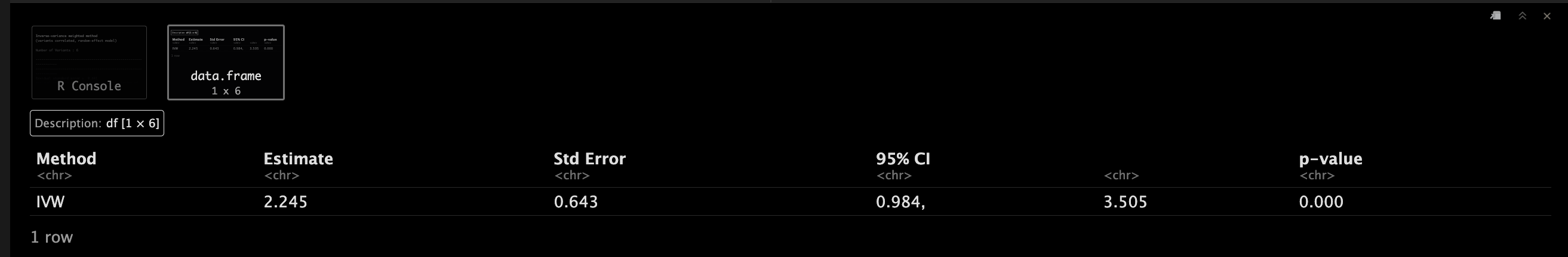
IVWObject.correl <- mr_ivw(MRInputObject.cor,
model = "default",
correl = TRUE,
distribution = "normal",
alpha = 0.05)
IVWObject.correl <- mr_ivw(mr_input(bx = calcium, bxse = calciumse,
by = fastgluc, byse = fastglucse, corr = calc.rho)
)
IVWObject.correl
5方法二 Median-based method
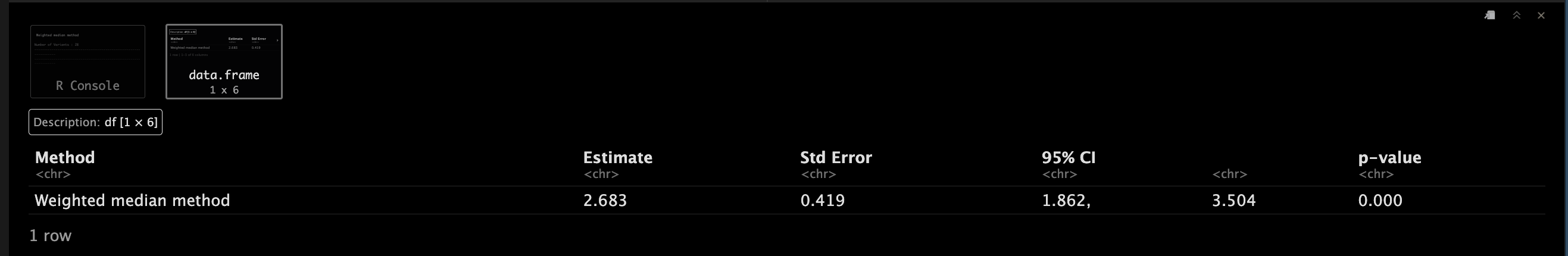
WeightedMedianObject <- mr_median(MRInputObject,
weighting = "weighted",
distribution = "normal",
alpha = 0.05,
iterations = 10000,
seed = 314159265)
WeightedMedianObject <- mr_median(mr_input(bx = ldlc,
bxse = ldlcse,
by = chdlodds,
byse = chdloddsse))
WeightedMedianObject
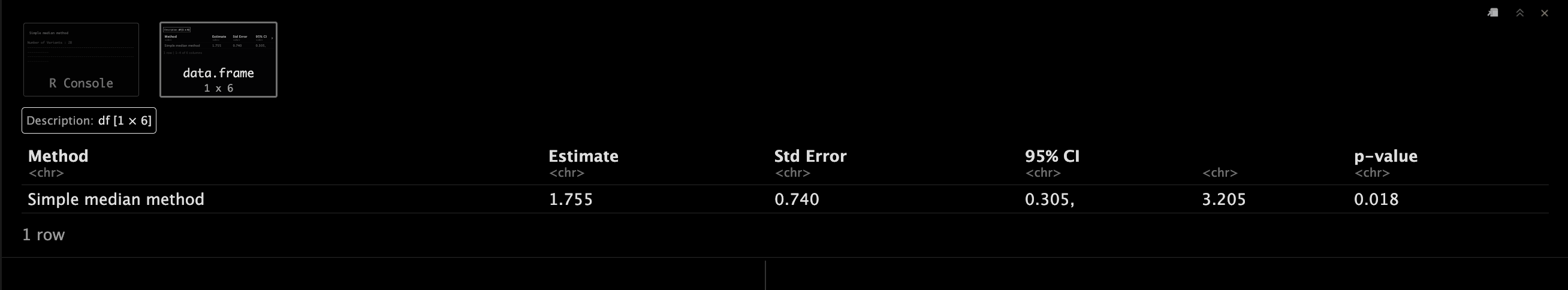
SimpleMedianObject <- mr_median(mr_input(bx = ldlc,
bxse = ldlcse,
by = chdlodds,
byse = chdloddsse),
weighting = "simple")
SimpleMedianObject
6方法三 MR-Egger method
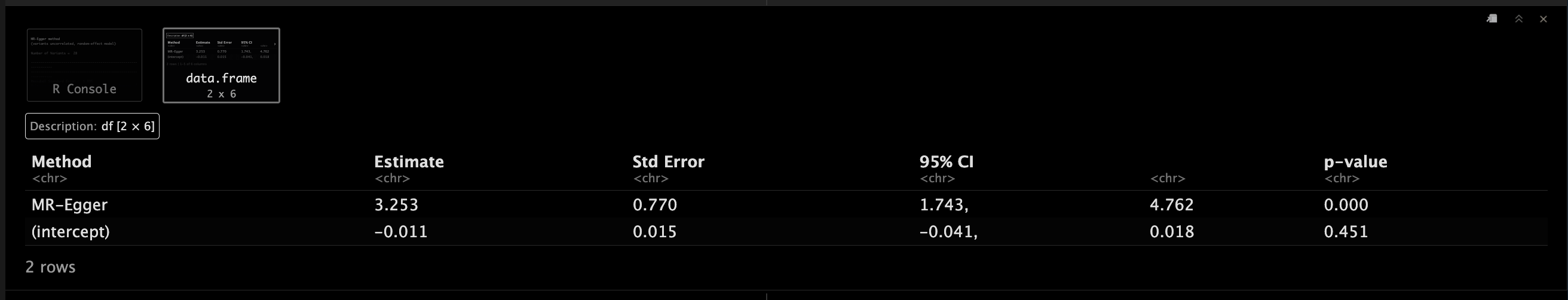
EggerObject <- mr_egger(MRInputObject,
robust = FALSE,
penalized = FALSE,
correl = FALSE,
distribution = "normal",
alpha = 0.05)
EggerObject <- mr_egger(mr_input(bx = ldlc,
bxse = ldlcse,
by = chdlodds,
byse = chdloddsse)
)
EggerObject
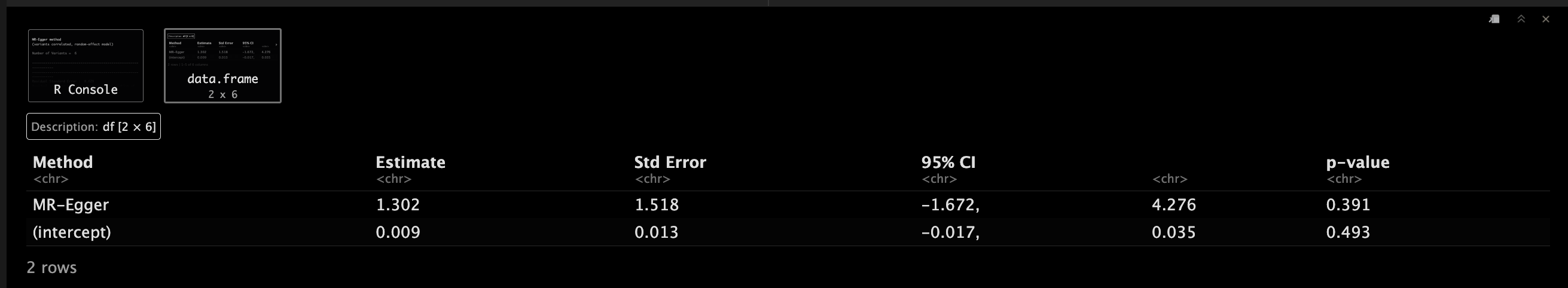
EggerObject.corr <- mr_egger(MRInputObject.cor,
correl = TRUE,
distribution = "normal",
alpha = 0.05)
EggerObject.corr <- mr_egger(mr_input(bx = calcium,
bxse = calciumse,
by = fastgluc,
byse = fastglucse,
corr = calc.rho))
EggerObject.corr
7方法四 Maximum likelihood method
MaxLikObject <- mr_maxlik(MRInputObject,
model = "default",
correl = FALSE,
psi = 0,
distribution = "normal",
alpha = 0.05)
MaxLikObject <- mr_maxlik(mr_input(bx = ldlc, bxse = ldlcse,by = chdlodds, byse = chdloddsse))
MaxLikObject
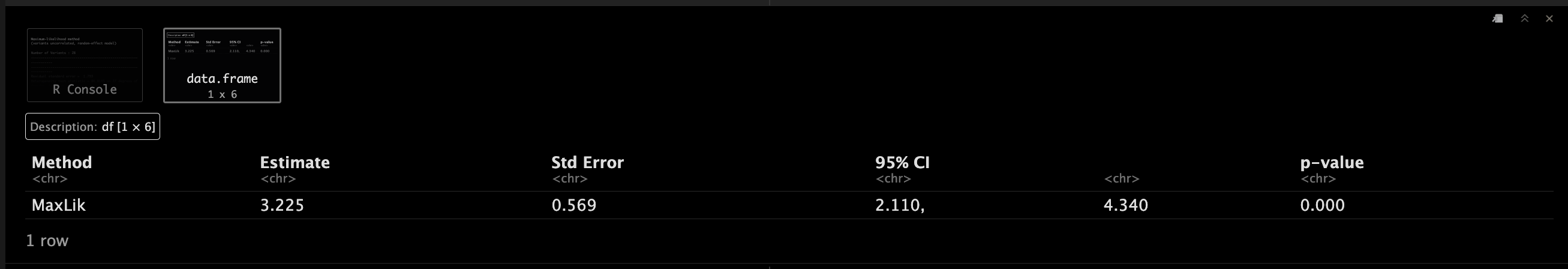
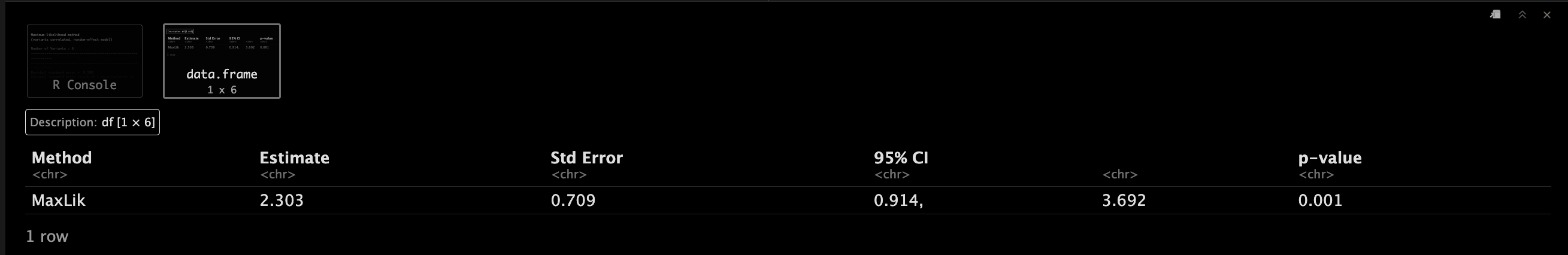
MaxLikObject.corr <- mr_maxlik(mr_input(bx = calcium, bxse = calciumse,by = fastgluc, byse = fastglucse, corr = calc.rho))
MaxLikObject.corr
8简单可视化
这里就以IVW为例哦。🙃
mr_plot(MRInputObject,
error = T,
orientate = F,
line = "ivw", # "ivw", "egger"
interactive = F,
labels = T
)


点个在看吧各位~ ✐.ɴɪᴄᴇ ᴅᴀʏ 〰
📍 🤩 LASSO | 不来看看怎么美化你的LASSO结果吗!?
📍 🤣 chatPDF | 别再自己读文献了!让chatGPT来帮你读吧!~
📍 🤩 WGCNA | 值得你深入学习的生信分析方法!~
📍 🤩 ComplexHeatmap | 颜狗写的高颜值热图代码!
📍 🤥 ComplexHeatmap | 你的热图注释还挤在一起看不清吗!?
📍 🤨 Google | 谷歌翻译崩了我们怎么办!?(附完美解决方案)
📍 🤩 scRNA-seq | 吐血整理的单细胞入门教程
📍 🤣 NetworkD3 | 让我们一起画个动态的桑基图吧~
📍 🤩 RColorBrewer | 再多的配色也能轻松搞定!~
📍 🧐 rms | 批量完成你的线性回归
📍 🤩 CMplot | 完美复刻Nature上的曼哈顿图
📍 🤠 Network | 高颜值动态网络可视化工具
📍 🤗 boxjitter | 完美复刻Nature上的高颜值统计图
📍 🤫 linkET | 完美解决ggcor安装失败方案(附教程)
📍 ......




本文由 mdnice 多平台发布




















 6463
6463











 被折叠的 条评论
为什么被折叠?
被折叠的 条评论
为什么被折叠?








