文章目录
前言
本课程来自深度之眼,部分截图来自课程视频。
文章标题:Neural Message Passing for Quantum Chemistry
神经网络消息传递应用量子化学(MPNN)
作者:Justin Gilmer, Samuel S. Schoenholz, Patrick F. Riley, Oriol Vinyals, George E. Dahl
单位:Google Brain
发表会议及时间:ICML 2017
公式输入请参考:在线Latex公式
graph isomorphism:图的同构性,指无论图的节点输入模型的先后顺序如何变化,图节点的embedding可以保持不变。
论文结构
- Abstract:提出化学分子上的监督学习具有研究意义。提出了一个统一的消息传递和信息汇合的框架。使用MPNN这个框架在化学分子性质预测任务上验证了模型的有效性。
- Introduction:回顾化学分子预测基于传统特征工程的算法,介绍将图神经网络模型引入,用MPNN框架在QM9数据集上评估模型。
- Message Passing Neural Networks:将GGNN、GCN等模型归入到统一的MPNN框架。
- Related Work:回顾化学分子预测的方法如DFT、KRR等算法。
- QM9 Dataset:介绍QM9数据集。
- MPNN Variants:讨论MPNN一些模型变体,如考虑边的特征、virtual边、master点、multiple towers策略等。
- Input Representation:QM9数据集的建图以及特征计算。
- Training&Results:模型参数设置以及实验效果展示,预测准确率、spatial信息、towers策略等。
- Conclusions and Future Work:提出了图神经网络一个统一的框架,消息的传递和汇合,以及图表征。
学习目标
泛读
背景
这十篇文章的目标都一样,就是要学习到节点的embedding表示(当然还有学习到整个图的表征的),就是下图中的第一个箭头,有了节点的embedding表征之后,就可以进行一些下游任务,例如预测邻居节点,预测节点的属性(标签等),目前对如何得到节点表征研究以及比较多了,可以看看如何针对下游任务设计具体的GNN应用。

回顾GNN的应用有很多方面,但凡是有以下结构都可以用到GNN:

对于本文而言,主要是针对化学的分子式来进行研究
例如,可把下面一个个化学分子式看做是一个个图,然后对其标签进行监督学习。

- Current methods
Use off-the-shelf fingerprint software to generate fixed-len feature vector for an arbitrary sized molecule.这里的用专门的软件进行特征工程,然后生成相应的分子式
Feed the generated feature vector to a neural network.然后丢入NN或者其他分类模型中,完成上面提到的分类任务。 - GNN advantages
Predictive performance: Machine-optimized fingerprints have better predictive performance than fixed fingerprints.这里的fingerprints 可以理解为embedding。GNN效果要比特征工程的结果好
Parsimony: Fixed fingerprints must be extremely large to encode all possible substructures without overlap. Neural fingerprints can be optimized to encode only relevant features, reducing downstream computation and regularization requirements.特征工程需要大量的先验知识,比较麻烦
Interpretability: No notion of similarity in fixed fingerprints. Neural fingerprint feature can be activated by similar but distinct molecular fragments.NN模型产生的embedding有一定的可解释性(这里是指分类后的空间距离是可以分开的)
模型框架
主要是消息传递机制的两个步骤,具体的可以看下GAT的讲解,这里直接贴图,不解释了:
·Step 1:Gather and transform the messages from neighbors:
AGG是汇聚函数,可以是求和/平均/LSTM等

·Step 2:Update the state of the target node.

用图表示:

本文模型框架
Message function:
m
v
t
+
1
=
∑
w
∈
N
v
M
(
h
v
t
,
h
w
t
,
e
v
w
)
m_v^{t+1}=\sum_{w\in N_v}M(h_v^t,h_w^t,e_{vw})
mvt+1=w∈Nv∑M(hvt,hwt,evw)
Update function:
h
v
t
+
1
=
U
(
h
v
t
,
m
v
t
+
1
)
h_v^{t+1}=U(h_v^t,m_v^{t+1})
hvt+1=U(hvt,mvt+1)
Readout function:
y
^
=
R
(
{
h
v
T
∣
v
∈
G
}
)
\hat y=R(\{h_v^T|v\in G\})
y^=R({hvT∣v∈G})
假如有如下图结构:

图中边的类型是不一样的,假如要对点v做消息汇聚操作。
对于点粉色点和橙色点来说,二者的边是实线:


看粉色和灰色点:


粉色红色:

粉色蓝色:

最后得到邻居的信息然后汇聚起来:

然后是框架的第二步:

用第t步的节点embedding和汇聚得到的消息,用更新函数得到黄色点
然后对图中所有节点都做上面两步:

经过T个时间步后,得到图所有节点的embedding后,可以进行readout

readout 函数例子:
R
=
f
(
∑
v
h
v
t
)
R=f(\sum_vh_v^t)
R=f(v∑hvt)
This readout function sums the current hidden states of all the nodes and computes an output through a learnable neural network
f
f
f
研究意义
• 提出图神经网络的一个统一框架MPNN
• 化学分子预测任务达到效果最优
• 提出了一些方法,包括virtual edges, master node, multiple towers等trick
• 将MPNN模型应用于化学分子任务并设计相应模型
摘要
- 化学分子的有监督学习具有巨大的潜力,可用于化学,药物发现和材料科学。
Supervised learning on molecules has incredible potential to be useful in chemistry, drug discovery, and materials science. - 之前的模型学习消息传递和聚合过程,并得到整个图的输出表征。
These models learn a message passing algorithm and aggregation procedure to compute a function of their entire input graph. - 在本论文中,我们将现有模型重新统一构建为消息传递神经网络(MPNN)通用框架,并探索该框架内的其他新颖变体应用于化学分子预测。
In this paper, we reformulate existing models into a single common framework we call Message Passing Neural Networks (MPNNs) and explore additional novel variations within this framework. - 使用MPNN框架,本文在重要的化学分子性能预测标准上证明了模型的有效性。
Using MPNNs we demonstrate state of the art results on an important molecular property prediction benchmark; these results are strong enough that we believe future work should focus on datasets with larger molecules or more accurate ground truth labels.
论文标题
- Introduction
- Message Passing Neural Networks
2.1Moving Forward - Related Work
- QM9 Dataset
- MPNN Variants
5.1Message Functions
5.2 Virtual Graph Elements
5.3 Readout Functions
5.4Multiple Towers - Input Representation
- Training
- Results
- Conclusions and Future Work
- Appendix
10.1Interpretation of Laplacian based models as MPNNs
10.2 A More Detailed Description of the Quantum Properties
10.3 Chemical Accuracy and DFT Error
10.4 Additional Results
模型精讲


任务简介:化学分子预测

从上图中可以看到MPNN模型比传统的特征工程速度上要快很多很多。
细节一:MPNN框架
Message function:
m
v
t
+
1
=
∑
w
∈
N
v
M
(
h
v
t
,
h
w
t
,
e
v
w
)
m_v^{t+1}=\sum_{w\in N_v}M(h_v^t,h_w^t,e_{vw})
mvt+1=w∈Nv∑M(hvt,hwt,evw)
这里要注意一下,原文在将MPNN框架套用在其他GNN模型的时候,上式的求和号没有表示出来,只求了单个邻居节点的情况。
Update function:
h
v
t
+
1
=
U
(
h
v
t
,
m
v
t
+
1
)
h_v^{t+1}=U(h_v^t,m_v^{t+1})
hvt+1=U(hvt,mvt+1)
Readout function:
y
^
=
R
(
{
h
v
T
∣
v
∈
G
}
)
\hat y=R(\{h_v^T|v\in G\})
y^=R({hvT∣v∈G})
MUR是模型要学习的三个参数,对应原文第二节。
For simplicity we describe MPNNs which operate on undirected graphs G with node features
x
v
x_v
xv and edge features
e
v
w
e_{vw}
evw. It is trivial to extend the formalism to directed multigraphs.
MPNN里面用的是无向图,由于可以处理不同类型的边,因此它也可以处理有向图(把不同指向的边看成不同类型)
The forward pass has two phases, a message passing phase and a readout phase. The message passing phase runs for
T
T
T time steps and is defined in terms of message functions
M
t
M_t
Mt and vertex update functions
U
t
U_t
Ut.
这里是把汇聚+更新做一个step,readout做一个step,通常我们是把汇聚和更新各看做一个step,这里的T是图网的层数。
框架中边的embedding(
h
e
v
w
t
h_{e_{vw}}^t
hevwt)也是可以学习到的。
细节二:用MPNN理解的模型
套用这个框架,可以看一下GNN的文章:
Duvenaud et al. Convolutional networks on graphs for learning molecular fingerprints. NIPS 2015.
里面提到的核心算法为:

其中第一行R代表GNN的层数,H是每一层要学习的参数。
第二行代表使用全0初始化f
第三行中提到的a是分子中的每一个原子,这里可以理解为图中的某一个节点
第四行代表得到该原子的特征作为GNN的输入
第五行代表循环R次(有R层,相当于对每一层都做以下操作)
第六行是对分子式中的每一个节点a进行循环
第七话代表是对a的所有邻居的特征r
第八行是对所有邻居特征进行汇聚操作,这里用的是sum
第九行是汇聚结果乘上第L层的GNN参数H,然后通过非线性变换得到当前节点的embedding,相当于本文的update function
第10/11行是GGNN的readout操作,得到整个图的embedding:f
MPNN代表模型还有:
Gated Graph Neural Networks (GG-NN): GRU as update function U.这个模型我们学过,它的readout直接用的是第T层的节点embedding来求的
Interaction Networks: an external vector representing some outside influence on the vertex v.这个算法在update function里面额外考虑了一个作用在点v的向量
x
v
x_v
xv,它的update function长成这个样子:
U
(
h
v
,
x
v
,
m
v
)
U(h_v,x_v,m_v)
U(hv,xv,mv)
上面的三个玩意是concatenate在一起的。
Molecular Graph Convolutions: updated edge representations.
这个算法有一个缺点,就是分别对点和边的信息进行汇聚,没有将二者结合起来考虑,原文说这样做:unable to identify correlations between edge states and node states.
Deep Tensor Neural Networks:/
Laplacian Based Methods: GCN. et al
细节三:MPNN化学分子预测
Start from GGNN model: Edge Network; Pair Message Strategies:
·Virtual Graph Elements: Virtual edge type; master node(远距离信息的传输)
·Readout Functions: set2set(不受顺序影响)
·Efficiency: multiple towers(类似多头注意力机制)
Edge Network
将GGNN中的汇聚函数形式为:
M
(
h
v
,
h
w
,
e
v
w
)
=
A
e
v
w
h
w
M(h_v,h_w,e_{vw})=A_{e_{vw}}h_w
M(hv,hw,evw)=Aevwhw
这个论文里面把按边设置的邻接矩阵
A
e
v
w
A_{e_{vw}}
Aevw替换为参数A乘以边特征的
e
v
w
e_{vw}
evw的形式:
M
(
h
v
,
h
w
,
e
v
w
)
=
A
(
e
v
w
)
h
w
M(h_v,h_w,e_{vw})=A(e_{vw})h_w
M(hv,hw,evw)=A(evw)hw
这里参数A是d×d×d维的,
e
v
w
e_{vw}
evw是d×1维的,
A
e
v
w
Ae_{vw}
Aevw是d×d维的,
h
w
h_w
hw是d×1维的,整个结果是d维的
也就是多了一个参数,可以学习到边的embedding。
Pair Message
从上面的结果可以看到,结果里面只考虑w这么一个点,但是边e是连接两个节点的,所以这里要把另外一个节点v也考虑进来,所以,汇聚函数就写成了:
m
v
w
=
f
(
h
w
t
,
h
v
t
,
e
v
w
)
m_{vw}=f(h_w^t,h_v^t,e_{vw})
mvw=f(hwt,hvt,evw)
其中
f
f
f是一个NN
对于有向图,可以分别计算
M
i
n
M^{in}
Min和
M
o
u
t
M^{out}
Mout
Virtual edge type+Master node
为不相连的节点添加虚拟边,使得节点能够相互连接,因此消息可以传递很远。
同样的,还为图添加主节点,该节点连接图中所有的节点(用的是特殊的边),且有一个单独的embedding
d
m
a
s
t
e
r
d_{master}
dmaster
Multiple towers
这个trick是针对dense graph而言,就是说考虑图中所有点两两相互连接,那么这个时候的边的数量e=n(n-1)/2,如果是在时间复杂度表示中可以表示为
n
2
n^2
n2,这个时候将d维的节点embedding(
h
v
t
h_v^t
hvt)分成k份,其维度就变成了
d
/
k
d/k
d/k维,embedding可以写为
h
v
t
,
k
h_v^{t,k}
hvt,k,然后每个部分分别计算消息的传递,得到下一个时间步的小份embedding:
h
v
t
+
1
,
k
,
v
∈
G
h_v^{t+1,k},v\in G
hvt+1,k,v∈G
理论上的时间复杂度从
O
(
n
2
d
2
)
O(n^2d^2)
O(n2d2)变成了
k
O
(
n
2
(
d
/
k
)
2
)
=
O
(
n
2
d
2
/
k
)
kO(n^2(d/k)^2)=O(n^2d^2/k)
kO(n2(d/k)2)=O(n2d2/k)
根据原文的描述,这k份小的embedding丢进了FC,使得这k份embedding相互关联:
(
h
v
t
,
1
,
h
v
t
,
2
,
⋯
,
h
v
t
,
k
)
=
g
(
h
~
v
t
,
1
,
h
~
v
t
,
2
,
⋯
,
h
~
v
t
,
k
)
(h_v^{t,1},h_v^{t,2},\cdots,h_v^{t,k})=g(\tilde h_v^{t,1},\tilde h_v^{t,2},\cdots,\tilde h_v^{t,k})
(hvt,1,hvt,2,⋯,hvt,k)=g(h~vt,1,h~vt,2,⋯,h~vt,k)
细节四:set2set模型
本文提到下面这个文章,如何得到一个序列集合的embedding。Oriol 也是本文作者之一。
Oriol Vinyals et al. Order Matters:Sequence to sequence for sets. ICLR 2016.


公式4.5.6是计算attention的。这三个公式可以看到是和集合中的元素顺序是无关的。
公式7是concat操作,然后结果又进入3里面的LSTM,所以经过的时间步越多,
q
t
∗
q^*_t
qt∗的维度越大。
set2set算法的目标是:
An important invariance property that must be satisfied when the input is a set (i.e., the order does not matter) is that swapping two elements
x
i
x_i
xi and
x
j
x_j
xj in the set
X
X
X should not alter its encoding.
对于传统的算法而言,图表征用节点表征求和得来有点不靠谱,因为这样得到的图表征维度和节点表征一样,明显图要包含更多的信息,需要更大的维度才行。
细节五:专题总结

Survey:
A Comprehensive Survey on Graph Neural Networks
Deep Learning on Graphs:A Survey
Graph Neural Networks:A Review of Methods and Applications
Book:Introduction to Graph Neural Networks.
Zhiyuan Liu,Jie Zhou
最新的论文:NIPS,ICML,ICLR,KDD,SIGIR,AAAI,JCAl
实验结果及分析

一个化学分子可以有多个label,可以为每一个label单独训练一个模型,也可以所有label训练一个模型
表中的数字是越小越好。

上表显示了论文提到的几个trick都非常有效
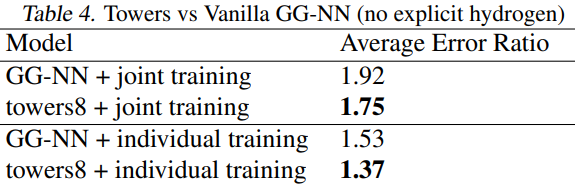
上表显示了towers的效果
joint training表示多个标签同时训练
individual training表示单个标签进行训练
论文总结
关键点
MPNN框架
多种GNN模型统一到MPNN框架
spatial/towers等技术
创新点
统一的消息传递框架
化学分子预测任务效果好
实证研究
启发点
提出统一的框架:MPNN
空域卷积分解为两个过程:消息传递与状态更新操作
将多个GNN模型如GGNN,GCN等按框架形式表达,简洁看到本质应用驱动型的创新和工作
GNN其他统一框架还有GN(Graph Network)、NLNN(non-local neural network)
复现

原文github
https://github.com/priba/nmp_qc
·python 3.7.6
·torch 1.3.1
·numpy 1.18.1
·networkx 2.4
·rdkit 2020.03.4对qm9建图
·jupyter notebook
pip install wget
pip install tensorboard _logger
conda install-c conda-forge rdkit
数据下载
python download.py qm9 -p ./
python download.py qm9 mutag enzymes
或者:
qm9数据集
https://github.com/priba/nmp_gc/tree/master/data
https://www.kaggle.com/zaharch/quantum-machine-9-aka-qm9
main.py
#!/usr/bin/python
# -*- coding: utf-8 -*-
"""
Trains a Neural Message Passing Model on various datasets. Methodologi defined in:
Gilmer, J., Schoenholz S.S., Riley, P.F., Vinyals, O., Dahl, G.E. (2017)
Neural Message Passing for Quantum Chemistry.
arXiv preprint arXiv:1704.01212 [cs.LG]
"""
# Torch
import torch
import torch.optim as optim
import torch.nn as nn
from torch.autograd import Variable
import time
import argparse
import os
import numpy as np
# Our Modules
import datasets
from datasets import utils
from models.MPNN import MPNN
from LogMetric import AverageMeter, Logger
__author__ = "Pau Riba, Anjan Dutta"
__email__ = "priba@cvc.uab.cat, adutta@cvc.uab.cat"
# Parser check
def restricted_float(x, inter):
x = float(x)
if x < inter[0] or x > inter[1]:
raise argparse.ArgumentTypeError("%r not in range [1e-5, 1e-4]"%(x,))
return x
# Argument parser
parser = argparse.ArgumentParser(description='Neural message passing')
#数据集
parser.add_argument('--dataset', default='qm9', help='QM9')
#数据集路径
parser.add_argument('--datasetPath', default='./data/qm9/dsgdb9nsd/', help='dataset path')
#1og记录路径
parser.add_argument('--logPath', default='./log/qm9/mpnn/', help='log path')
#画图
parser.add_argument('--plotLr', default=False, help='allow plotting the data')
#画图路径
parser.add_argument('--plotPath', default='./plot/qm9/mpnn/', help='plot path')
#checkpoint容错路径
parser.add_argument('--resume', default='./checkpoint/qm9/mpnn/',
help='path to latest checkpoint')
# Optimization Options
# batch size大小
parser.add_argument('--batch-size', type=int, default=100, metavar='N',
help='Input batch size for training (default: 20)')
#是否使用cuda
parser.add_argument('--no-cuda', action='store_true', default=False,
help='Enables CUDA training')
#epochs数量
parser.add_argument('--epochs', type=int, default=5, metavar='N',#默认要跑360次,这里跑个5次意思一下
help='Number of epochs to train (default: 360)')
# parser.add_argument('--epochs', type=int, default=360, metavar='N',
# help='Number of epochs to train (default: 360)')
#自适应的学习率
parser.add_argument('--lr', type=lambda x: restricted_float(x, [1e-5, 1e-2]), default=1e-3, metavar='LR',
help='Initial learning rate [1e-5, 5e-4] (default: 1e-4)')
#衰减系数
parser.add_argument('--lr-decay', type=lambda x: restricted_float(x, [.01, 1]), default=0.6, metavar='LR-DECAY',
help='Learning rate decay factor [.01, 1] (default: 0.6)')
#多少次epoch开始衰减
parser.add_argument('--schedule', type=list, default=[0.1, 0.9], metavar='S',
help='Percentage of epochs to start the learning rate decay [0, 1] (default: [0.1, 0.9])')
#momentum优化
parser.add_argument('--momentum', type=float, default=0.9, metavar='M',
help='SGD momentum (default: 0.9)')
# i/o,多少次epoch记录一次log
parser.add_argument('--log-interval', type=int, default=20, metavar='N',
help='How many batches to wait before logging training status')
# Accelerating
parser.add_argument('--prefetch', type=int, default=2, help='Pre-fetching threads.')
best_er1 = 0
def main():
global args, best_er1
args = parser.parse_args()
#jupyter
#args = parser.parse_args([])
# Check if CUDA is enabled
args.cuda = not args.no_cuda and torch.cuda.is_available()
# Load data
root = args.datasetPath
print('Prepare files')
#遍历root路径下所有的文件和文件夹,如果是文件则存到list
files = [f for f in os.listdir(root) if os.path.isfile(os.path.join(root, f))]
#打乱数据顺序
idx = np.random.permutation(len(files))
idx = idx.tolist()
# 原文的数据集划分,改小后是150个分子式
# valid_ids = [files[i] for i in idx[0:10000]]
# test_ids = [files[i] for i in idx[10000:20000]]
# train_ids = [files[i] for i in idx[20000:]]
# 打乱后的数据进行划分
valid_ids = [files[i] for i in idx[0:50]]
test_ids = [files[i] for i in idx[50:100]]
train_ids = [files[i] for i in idx[100:]]
data_train = datasets.Qm9(root, train_ids, edge_transform=utils.qm9_edges, e_representation='raw_distance')
data_valid = datasets.Qm9(root, valid_ids, edge_transform=utils.qm9_edges, e_representation='raw_distance')
data_test = datasets.Qm9(root, test_ids, edge_transform=utils.qm9_edges, e_representation='raw_distance')
# Define model and optimizer
print('Define model')
# Select one graph
g_tuple, l = data_train[0]
g, h_t, e = g_tuple
print('\tStatistics')
#get_graph_stats在utils里面,得到均值和标准差
stat_dict = datasets.utils.get_graph_stats(data_valid, ['target_mean', 'target_std'])
#三个数据集归一化
#data_norm = (data-mean)/std
data_train.set_target_transform(lambda x: datasets.utils.normalize_data(x,stat_dict['target_mean'],
stat_dict['target_std']))
data_valid.set_target_transform(lambda x: datasets.utils.normalize_data(x, stat_dict['target_mean'],
stat_dict['target_std']))
data_test.set_target_transform(lambda x: datasets.utils.normalize_data(x, stat_dict['target_mean'],
stat_dict['target_std']))
# Data Loader
#封装成DataLoader
#注意这里面的collate_g函数,在utils里面
train_loader = torch.utils.data.DataLoader(data_train,
batch_size=args.batch_size, shuffle=True,
collate_fn=datasets.utils.collate_g,
num_workers=args.prefetch, pin_memory=True)
valid_loader = torch.utils.data.DataLoader(data_valid,
batch_size=args.batch_size, collate_fn=datasets.utils.collate_g,
num_workers=args.prefetch, pin_memory=True)
test_loader = torch.utils.data.DataLoader(data_test,
batch_size=args.batch_size, collate_fn=datasets.utils.collate_g,
num_workers=args.prefetch, pin_memory=True)
print('\tCreate model')
#点的特征维度,边的特征维度
in_n = [len(h_t[0]), len(list(e.values())[0])]
#hidden state/embedding维度
hidden_state_size = 73
#邻居消息m_i维度(聚合后的维度)后面都用d_v表示
message_size = 73
#GNN层数
n_layers = 3
#labels数量
l_target = len(l)
#回归任务
type ='regression'
#定义mpnn模型
model = MPNN(in_n, hidden_state_size, message_size, n_layers, l_target, type=type)
del in_n, hidden_state_size, message_size, n_layers, l_target, type
print('Optimizer')
optimizer = optim.Adam(model.parameters(), lr=args.lr)
#回归任务使用MSE 1oss
criterion = nn.MSELoss()
#评估指标,|a-b|/|b|
evaluation = lambda output, target: torch.mean(torch.abs(output - target) / torch.abs(target))
print('Logger')
#日志记录
logger = Logger(args.logPath)
lr_step = (args.lr-args.lr*args.lr_decay)/(args.epochs*args.schedule[1] - args.epochs*args.schedule[0])
# get the best checkpoint if available without training
if args.resume:
checkpoint_dir = args.resume
best_model_file = os.path.join(checkpoint_dir, 'model_best.pth')
if not os.path.isdir(checkpoint_dir):
os.makedirs(checkpoint_dir)
if os.path.isfile(best_model_file):
print("=> loading best model '{}'".format(best_model_file))
checkpoint = torch.load(best_model_file)
args.start_epoch = checkpoint['epoch']
best_acc1 = checkpoint['best_er1']
model.load_state_dict(checkpoint['state_dict'])
optimizer.load_state_dict(checkpoint['optimizer'])
print("=> loaded best model '{}' (epoch {})".format(best_model_file, checkpoint['epoch']))
else:
print("=> no best model found at '{}'".format(best_model_file))
print('Check cuda')
if args.cuda:
print('\t* Cuda')
model = model.cuda()
criterion = criterion.cuda()
# Epoch for loop
for epoch in range(0, args.epochs):
if epoch > args.epochs * args.schedule[0] and epoch < args.epochs * args.schedule[1]:
args.lr -= lr_step
for param_group in optimizer.param_groups:
param_group['lr'] = args.lr
# train for one epoch
train(train_loader, model, criterion, optimizer, epoch, evaluation, logger)
# evaluate on test set
er1 = validate(valid_loader, model, criterion, evaluation, logger)
is_best = er1 > best_er1
best_er1 = min(er1, best_er1)
utils.save_checkpoint({'epoch': epoch + 1, 'state_dict': model.state_dict(), 'best_er1': best_er1,
'optimizer': optimizer.state_dict(), }, is_best=is_best, directory=args.resume)
# Logger step
logger.log_value('learning_rate', args.lr).step()
# get the best checkpoint and test it with test set
if args.resume:
checkpoint_dir = args.resume
best_model_file = os.path.join(checkpoint_dir, 'model_best.pth')
if not os.path.isdir(checkpoint_dir):
os.makedirs(checkpoint_dir)
if os.path.isfile(best_model_file):
print("=> loading best model '{}'".format(best_model_file))
checkpoint = torch.load(best_model_file)
args.start_epoch = checkpoint['epoch']
best_acc1 = checkpoint['best_er1']
model.load_state_dict(checkpoint['state_dict'])
if args.cuda:
model.cuda()
optimizer.load_state_dict(checkpoint['optimizer'])
print("=> loaded best model '{}' (epoch {})".format(best_model_file, checkpoint['epoch']))
else:
print("=> no best model found at '{}'".format(best_model_file))
# For testing
validate(test_loader, model, criterion, evaluation)
def train(train_loader, model, criterion, optimizer, epoch, evaluation, logger):
batch_time = AverageMeter()
data_time = AverageMeter()
losses = AverageMeter()
error_ratio = AverageMeter()
# switch to train mode
model.train()
end = time.time()
for i, (g, h, e, target) in enumerate(train_loader):
# Prepare input data
if args.cuda:
g, h, e, target = g.cuda(), h.cuda(), e.cuda(), target.cuda()
g, h, e, target = Variable(g), Variable(h), Variable(e), Variable(target)
# Measure data loading time
data_time.update(time.time() - end)
optimizer.zero_grad()
# Compute output
output = model(g, h, e)
train_loss = criterion(output, target)
# Logs
# losses.update(train_loss.data[0], g.size(0))
losses.update(train_loss.data, g.size(0))
# error_ratio.update(evaluation(output, target).data[0], g.size(0))
error_ratio.update(evaluation(output, target).data, g.size(0))
# compute gradient and do SGD step
train_loss.backward()
optimizer.step()
# Measure elapsed time
batch_time.update(time.time() - end)
end = time.time()
if i % args.log_interval == 0 and i > 0:
print('Epoch: [{0}][{1}/{2}]\t'
'Time {batch_time.val:.3f} ({batch_time.avg:.3f})\t'
'Data {data_time.val:.3f} ({data_time.avg:.3f})\t'
'Loss {loss.val:.4f} ({loss.avg:.4f})\t'
'Error Ratio {err.val:.4f} ({err.avg:.4f})'
.format(epoch, i, len(train_loader), batch_time=batch_time,
data_time=data_time, loss=losses, err=error_ratio))
logger.log_value('train_epoch_loss', losses.avg)
logger.log_value('train_epoch_error_ratio', error_ratio.avg)
print('Epoch: [{0}] Avg Error Ratio {err.avg:.3f}; Average Loss {loss.avg:.3f}; Avg Time x Batch {b_time.avg:.3f}'
.format(epoch, err=error_ratio, loss=losses, b_time=batch_time))
def validate(val_loader, model, criterion, evaluation, logger=None):
batch_time = AverageMeter()
losses = AverageMeter()
error_ratio = AverageMeter()
# switch to evaluate mode
model.eval()
end = time.time()
for i, (g, h, e, target) in enumerate(val_loader):
# Prepare input data
if args.cuda:
g, h, e, target = g.cuda(), h.cuda(), e.cuda(), target.cuda()
g, h, e, target = Variable(g), Variable(h), Variable(e), Variable(target)
# Compute output
output = model(g, h, e)
# Logs
# losses.update(criterion(output, target).data[0], g.size(0))
# error_ratio.update(evaluation(output, target).data[0], g.size(0))
losses.update(criterion(output, target).data, g.size(0))
error_ratio.update(evaluation(output, target).data, g.size(0))
# measure elapsed time
batch_time.update(time.time() - end)
end = time.time()
if i % args.log_interval == 0 and i > 0:
print('Test: [{0}/{1}]\t'
'Time {batch_time.val:.3f} ({batch_time.avg:.3f})\t'
'Loss {loss.val:.4f} ({loss.avg:.4f})\t'
'Error Ratio {err.val:.4f} ({err.avg:.4f})'
.format(i, len(val_loader), batch_time=batch_time,
loss=losses, err=error_ratio))
print(' * Average Error Ratio {err.avg:.3f}; Average Loss {loss.avg:.3f}'
.format(err=error_ratio, loss=losses))
if logger is not None:
logger.log_value('test_epoch_loss', losses.avg)
logger.log_value('test_epoch_error_ratio', error_ratio.avg)
return error_ratio.avg
if __name__ == '__main__':
main()
qm9
#!/usr/bin/python
# -*- coding: utf-8 -*-
"""
qm9.py:
Usage:
"""
# Networkx should be imported before torch
import networkx as nx
import torch.utils.data as data
import numpy as np
import argparse
import datasets.utils as utils
import time
import os,sys
import torch
reader_folder = os.path.realpath( os.path.abspath('../GraphReader'))
if reader_folder not in sys.path:
sys.path.insert(1, reader_folder)
from GraphReader.graph_reader import xyz_graph_reader
__author__ = "Pau Riba, Anjan Dutta"
__email__ = "priba@cvc.uab.cat, adutta@cvc.uab.cat"
class Qm9(data.Dataset):
# Constructor
def __init__(self, root_path, ids, vertex_transform=utils.qm9_nodes, edge_transform=utils.qm9_edges,
target_transform=None, e_representation='raw_distance'):
#路径
self.root = root_path
#index
self.ids = ids
#未指定,默认值utils.qm9_nodes
self.vertex_transform = vertex_transform
#utils.qm9_edges
self.edge_transform = edge_transform
#未指定,默认值None
self.target_transform = target_transform
#化学分子距离度量方式,有三种,这里用的是'raw_distance'
self.e_representation = e_representation
def __getitem__(self, index):
#读图,返回networkx类型的图,标签list
#self.ids[index]是根据index得到化学分子文件名
#self.root是路径
g, target = xyz_graph_reader(os.path.join(self.root, self.ids[index]))
#返回h每个点的特征(list of list),在utils里面
if self.vertex_transform is not None:
h = self.vertex_transform(g)
#返回g邻接矩阵;e是词典,key是边,value是特征,在utils里面
if self.edge_transform is not None:
g, e = self.edge_transform(g, self.e_representation)
#这里是不用执行
if self.target_transform is not None:
target = self.target_transform(target)
#g:邻接矩阵
#h:每个点的特征(list of list)
#e:词典,key是边,value是特征
#target:标签list
return (g, h, e), target
def __len__(self):
return len(self.ids)
def set_target_transform(self, target_transform):
self.target_transform = target_transform
if __name__ == '__main__':
# Parse optios for downloading
parser = argparse.ArgumentParser(description='QM9 Object.')
# Optional argument
parser.add_argument('--root', nargs=1, help='Specify the data directory.', default=['../data/qm9/dsgdb9nsd'])
args = parser.parse_args()
root = args.root[0]
files = [f for f in os.listdir(root) if os.path.isfile(os.path.join(root, f))]
idx = np.random.permutation(len(files))
idx = idx.tolist()
valid_ids = [files[i] for i in idx[0:10000]]
test_ids = [files[i] for i in idx[10000:20000]]
train_ids = [files[i] for i in idx[20000:]]
data_train = Qm9(root, train_ids, vertex_transform=utils.qm9_nodes, edge_transform=lambda g: utils.qm9_edges(g, e_representation='raw_distance'))
data_valid = Qm9(root, valid_ids)
data_test = Qm9(root, test_ids)
print(len(data_train))
print(len(data_valid))
print(len(data_test))
print(data_train[1])
print(data_valid[1])
print(data_test[1])
start = time.time()
print(utils.get_graph_stats(data_valid, 'degrees'))
end = time.time()
print('Time Statistics Par')
print(end - start)
graph_reader
#!/usr/bin/python
# -*- coding: utf-8 -*-
"""
graph_reader.py: Reads graph datasets.
Usage:
"""
import numpy as np
import networkx as nx
import random
import argparse
from rdkit import Chem
from rdkit.Chem import ChemicalFeatures
from rdkit import RDConfig
import os
from os import listdir
from os.path import isfile, join
import xml.etree.ElementTree as ET
__author__ = "Pau Riba, Anjan Dutta"
__email__ = "priba@cvc.uab.cat, adutta@cvc.uab.cat"
random.seed(2)
np.random.seed(2)
def load_dataset(directory, dataset, subdir = '01_Keypoint' ):
if dataset == 'enzymes':
file_path = join(directory, dataset)
files = [f for f in listdir(file_path) if isfile(join(file_path, f))]
classes = []
graphs = []
for i in range(len(files)):
g, c = create_graph_enzymes(join(directory, dataset, files[i]))
graphs += [g]
classes += [c]
train_graphs, train_classes, valid_graphs, valid_classes, test_graphs, test_classes = divide_datasets(graphs, classes)
elif dataset == 'mutag':
file_path = join(directory, dataset)
files = [f for f in listdir(file_path) if isfile(join(file_path, f))]
classes = []
graphs = []
for i in range(len(files)):
g, c = create_graph_mutag(join(directory, dataset, files[i]))
graphs += [g]
classes += [c]
train_graphs, train_classes, valid_graphs, valid_classes, test_graphs, test_classes = divide_datasets(graphs, classes)
elif dataset == 'MUTAG' or dataset == 'ENZYMES' or dataset == 'NCI1' or \
dataset == 'NCI109' or dataset == 'DD':
label_file = dataset + '.label'
list_file = dataset + '.list'
label_file_path = join(directory, dataset, label_file)
list_file_path = join(directory, dataset, list_file)
with open(label_file_path, 'r') as f:
l = f.read()
classes = [int(s) for s in l.split() if s.isdigit()]
with open(list_file_path, 'r') as f:
files = f.read().splitlines()
graphs = load_graphml(join(directory, dataset), files)
train_graphs, train_classes, valid_graphs, valid_classes, test_graphs, test_classes = divide_datasets(graphs, classes)
elif dataset == 'gwhist':
train_classes, train_files = read_2cols_set_files(join(directory,'Set/Train.txt'))
test_classes, test_files = read_2cols_set_files(join(directory,'Set/Test.txt'))
valid_classes, valid_files = read_2cols_set_files(join(directory,'Set/Valid.txt'))
train_classes, valid_classes, test_classes = \
create_numeric_classes(train_classes, valid_classes, test_classes)
data_dir = join(directory, 'Data/Word_Graphs/01_Skew', subdir)
train_graphs = load_gwhist(data_dir, train_files)
valid_graphs = load_gwhist(data_dir, valid_files)
test_graphs = load_gwhist(data_dir, test_files)
elif dataset == 'qm9':
file_path = join(directory, dataset, subdir)
files = [f for f in listdir(file_path) if isfile(join(file_path, f))]
data_dir = join(directory, dataset, subdir)
graphs , labels = load_qm9(data_dir, files)
# TODO: Split into train, valid and test sets and class information
idx = np.random.permutation(len(labels))
valid_graphs = [graphs[i] for i in idx[0:10000]]
valid_classes = [labels[i] for i in idx[0:10000]]
test_graphs = [graphs[i] for i in idx[10000:20000]]
test_classes = [labels[i] for i in idx[10000:20000]]
train_graphs = [graphs[i] for i in idx[20000:]]
train_classes = [labels[i] for i in idx[20000:]]
return train_graphs, train_classes, valid_graphs, valid_classes, test_graphs, test_classes
def create_numeric_classes(train_classes, valid_classes, test_classes):
classes = train_classes + valid_classes + test_classes
uniq_classes = sorted(list(set(classes)))
train_classes_ = [0] * len(train_classes)
valid_classes_ = [0] * len(valid_classes)
test_classes_ = [0] * len(test_classes)
for ix in range(len(uniq_classes)):
idx = [i for i, c in enumerate(train_classes) if c == uniq_classes[ix]]
for i in idx:
train_classes_[i] = ix
idx = [i for i, c in enumerate(valid_classes) if c == uniq_classes[ix]]
for i in idx:
valid_classes_[i] = ix
idx = [i for i, c in enumerate(test_classes) if c == uniq_classes[ix]]
for i in idx:
test_classes_[i] = ix
return train_classes_, valid_classes_, test_classes_
def load_gwhist(data_dir, files):
graphs = []
for i in range(len(files)):
g = create_graph_gwhist(join(data_dir, files[i]))
graphs += [g]
return graphs
def load_graphml(data_dir, files):
graphs = []
for i in range(len(files)):
g = nx.read_graphml(join(data_dir,files[i]))
graphs += [g]
return graphs
def load_qm9(data_dir, files):
graphs = []
labels = []
for i in range(len(files)):
g , l = xyz_graph_reader(join(data_dir, files[i]))
graphs += [g]
labels.append(l)
return graphs, labels
def read_2cols_set_files(file):
f = open(file, 'r')
lines = f.read().splitlines()
f.close()
classes = []
files = []
for line in lines:
c, f = line.split(' ')[:2]
classes += [c]
files += [f + '.gxl']
return classes, files
def read_cxl(file):
files = []
classes = []
tree_cxl = ET.parse(file)
root_cxl = tree_cxl.getroot()
for f in root_cxl.iter('print'):
files += [f.get('file')]
classes += [f.get('class')]
return classes, files
def divide_datasets(graphs, classes):
uc = list(set(classes))
tr_idx = []
va_idx = []
te_idx = []
for c in uc:
idx = [i for i, x in enumerate(classes) if x == c]
tr_idx += sorted(np.random.choice(idx, int(0.8*len(idx)), replace=False))
va_idx += sorted(np.random.choice([x for x in idx if x not in tr_idx], int(0.1*len(idx)), replace=False))
te_idx += sorted(np.random.choice([x for x in idx if x not in tr_idx and x not in va_idx], int(0.1*len(idx)), replace=False))
train_graphs = [graphs[i] for i in tr_idx]
valid_graphs = [graphs[i] for i in va_idx]
test_graphs = [graphs[i] for i in te_idx]
train_classes = [classes[i] for i in tr_idx]
valid_classes = [classes[i] for i in va_idx]
test_classes = [classes[i] for i in te_idx]
return train_graphs, train_classes, valid_graphs, valid_classes, test_graphs, test_classes
def create_graph_enzymes(file):
f = open(file, 'r')
lines = f.read().splitlines()
f.close()
# get the indices of the vertext, adj list and class
idx_vertex = lines.index("#v - vertex labels")
idx_adj_list = lines.index("#a - adjacency list")
idx_clss = lines.index("#c - Class")
# node label
vl = [int(ivl) for ivl in lines[idx_vertex+1:idx_adj_list]]
adj_list = lines[idx_adj_list+1:idx_clss]
sources = list(range(1,len(adj_list)+1))
for i in range(len(adj_list)):
if not adj_list[i]:
adj_list[i] = str(sources[i])
else:
adj_list[i] = str(sources[i])+","+adj_list[i]
g = nx.parse_adjlist(adj_list, nodetype=int, delimiter=",")
for i in range(1, g.number_of_nodes()+1):
g.nodes[i]['labels'] = np.array(vl[i-1])
c = int(lines[idx_clss+1])
return g, c
def create_graph_mutag(file):
f = open(file, 'r')
lines = f.read().splitlines()
f.close()
# get the indices of the vertext, adj list and class
idx_vertex = lines.index("#v - vertex labels")
idx_edge = lines.index("#e - edge labels")
idx_clss = lines.index("#c - Class")
# node label
vl = [int(ivl) for ivl in lines[idx_vertex+1:idx_edge]]
edge_list = lines[idx_edge+1:idx_clss]
g = nx.parse_edgelist(edge_list, nodetype=int, data=(('weight', float),), delimiter=",")
for i in range(1, g.number_of_nodes()+1):
g.nodes[i]['labels'] = np.array(vl[i-1])
c = int(lines[idx_clss+1])
return g, c
def create_graph_gwhist(file):
tree_gxl = ET.parse(file)
root_gxl = tree_gxl.getroot()
vl = []
for node in root_gxl.iter('node'):
for attr in node.iter('attr'):
if(attr.get('name') == 'x'):
x = attr.find('float').text
elif(attr.get('name') == 'y'):
y = attr.find('float').text
vl += [[x, y]]
g = nx.Graph()
for edge in root_gxl.iter('edge'):
s = edge.get('from')
s = int(s.split('_')[1])
t = edge.get('to')
t = int(t.split('_')[1])
g.add_edge(s, t)
for i in range(g.number_of_nodes()):
if i not in g.nodes:
g.add_node(i)
g.nodes[i]['labels'] = np.array(vl[i])
return g
def isfloat(value):
try:
float(value)
return True
except ValueError:
return False
def create_graph_grec(file):
tree_gxl = ET.parse(file)
root_gxl = tree_gxl.getroot()
vl = []
switch_node = {'circle': 0, 'corner': 1, 'endpoint': 2, 'intersection': 3}
switch_edge = {'arc': 0, 'arcarc': 1, 'line': 2, 'linearc': 3}
for node in root_gxl.iter('node'):
for attr in node.iter('attr'):
if (attr.get('name') == 'x'):
x = int(attr.find('Integer').text)
elif (attr.get('name') == 'y'):
y = int(attr.find('Integer').text)
elif (attr.get('name') == 'type'):
t = switch_node.get(attr.find('String').text, 4)
vl += [[x, y, t]]
g = nx.Graph()
for edge in root_gxl.iter('edge'):
s = int(edge.get('from'))
t = int(edge.get('to'))
for attr in edge.iter('attr'):
if(attr.get('name') == 'frequency'):
f = attr.find('Integer').text
elif(attr.get('name') == 'type0'):
ta = switch_edge.get(attr.find('String').text)
elif (attr.get('name') == 'angle0'):
a = attr.find('String').text
if isfloat(a):
a = float(a)
else:
a = 0.0 # TODO: The erroneous string is replaced with 0.0
g.add_edge(s, t, frequency=f, type=ta, angle=a)
for i in range(len(vl)):
if i not in g.nodes:
g.add_node(i)
g.nodes[i]['labels'] = np.array(vl[i][:3])
return g
def create_graph_letter(file):
tree_gxl = ET.parse(file)
root_gxl = tree_gxl.getroot()
vl = []
for node in root_gxl.iter('node'):
for attr in node.iter('attr'):
if (attr.get('name') == 'x'):
x = float(attr.find('float').text)
elif (attr.get('name') == 'y'):
y = float(attr.find('float').text)
vl += [[x, y]]
g = nx.Graph()
for edge in root_gxl.iter('edge'):
s = int(edge.get('from').split('_')[1])
t = int(edge.get('to').split('_')[1])
g.add_edge(s, t)
for i in range(len(vl)):
if i not in g.nodes:
g.add_node(i)
g.nodes[i]['labels'] = np.array(vl[i][:2])
return g
# Initialization of graph for QM9
def init_graph(prop):
#读取信息
prop = prop.split()
g_tag = prop[0]
g_index = int(prop[1])
g_A = float(prop[2])
g_B = float(prop[3])
g_C = float(prop[4])
#以下为labels
#论文section"4.QM9 Dataset"介绍
g_mu = float(prop[5])
g_alpha = float(prop[6])
g_homo = float(prop[7])
g_lumo = float(prop[8])
g_gap = float(prop[9])
g_r2 = float(prop[10])
g_zpve = float(prop[11])
g_U0 = float(prop[12])
g_U = float(prop[13])
g_H = float(prop[14])
g_G = float(prop[15])
g_Cv = float(prop[16])
#labels结束
labels = [g_mu, g_alpha, g_homo, g_lumo, g_gap, g_r2, g_zpve, g_U0, g_U, g_H, g_G, g_Cv]
#返回networkx类型的图,标签list
return nx.Graph(tag=g_tag, index=g_index, A=g_A, B=g_B, C=g_C, mu=g_mu, alpha=g_alpha, homo=g_homo,
lumo=g_lumo, gap=g_gap, r2=g_r2, zpve=g_zpve, U0=g_U0, U=g_U, H=g_H, G=g_G, Cv=g_Cv), labels
# XYZ file reader for QM9 dataset
def xyz_graph_reader(graph_file):
#数据例子:
# https://www.kaggle.com/zaharch/quantum-machine-9-aka-qm9?select=dsgdb9nsd_000001.xyz
'''
第一行 5
第二行 gdb 1 157.7118 157.70997 157.70699 0. 13.21 -0.3877 0.1171 0.5048 35.3641 0.044749 -40.47893 -40.476062 -40.475117 -40.498597 6.469
第三行 C -0.0126981359 1.0858041578 0.0080009958 -0.535689
第四行 H 0.002150416 -0.0060313176 0.0019761204 0.133921
第五行 H 1.0117308433 1.4637511618 0.0002765748 0.133922
第六行 H -0.540815069 1.4475266138 -0.8766437152 0.133923
第七行 H -0.5238136345 1.4379326443 0.9063972942 0.133923
第八行 1341.307 1341.3284 1341.365 1562.6731 1562.7453 3038.3205 3151.6034 3151.6788 3151.7078
第九行 C C
第十行 InChI=1S/CH4/h1H4 InChI=1S/CH4/h1H4
'''
with open(graph_file,'r') as f:
# Number of atoms
# 第一行原子数量,上例中是5
na = int(f.readline())
# Graph properties
#第二行,图的性质(ground truth labels)
properties = f.readline()
#得到networkx类型的图,内含标签list
g, l = init_graph(properties)
atom_properties = []
# Atoms properties
#根据第一行读到的原子数量na,读取na行得到每个原子的信息
for i in range(na):
a_properties = f.readline()#每一行都存为一个list,
a_properties = a_properties.replace('.*^', 'e')
a_properties = a_properties.replace('*^', 'e')
a_properties = a_properties.split()
atom_properties.append(a_properties)#atom_properties是list套list
# Frequencies
#这一行信息互略
f.readline()
# SMILES(Simplified molecular input line entry specification),简化分子线性输入规范,是一种用ASCII字符串明确描述分子结构的规范。
smiles = f.readline()
smiles = smiles.split()
smiles = smiles[0]
#参考rdkit包
# https://www.rdkit.org/docs/source/rdkit.Chem.html
#smiles转换为分子对象
# https://zhuanlan.zhihu.com/p/82497166
m = Chem.MolFromSmiles(smiles)
m = Chem.AddHs(m)#按原文加入氢原子
#from rdkit import RDConfig
#https://zhuanlan.zhihu.com/p/141908982
#导入特征库
fdef_name = os.path.join(RDConfig.RDDataDir, 'BaseFeatures.fdef')
#创建特征工厂
factory = ChemicalFeatures.BuildFeatureFactory(fdef_name)
#计算化学特征
feats = factory.GetFeaturesForMol(m)
# Create nodes
#读入点
for i in range(0, m.GetNumAtoms()):
# https://zhuanlan.zhihu.com/p/143111689
# 通过索引获取原子
atom_i = m.GetAtomWithIdx(i)
#获取原子符号:Getsymbol()
#获取原子序号:GetAtomicNum()
#获取原子杂化方式:GetHybridization()
# atom properties是上面处理的原子的信息
g.add_node(i, a_type=atom_i.GetSymbol(), a_num=atom_i.GetAtomicNum(), acceptor=0, donor=0,
aromatic=atom_i.GetIsAromatic(), hybridization=atom_i.GetHybridization(),
num_h=atom_i.GetTotalNumHs(), coord=np.array(atom_properties[i][1:4]).astype(np.float),
pc=float(atom_properties[i][4]))
# https://zhuanlan.zhihu.com/p/141908982
#遍历化学特征feats,找到包含供体、受体的原子的id(GetAtomIds()),把这些原子id放到node_list,然后遍历node_list,分别加donor和acceptor属性
#搜索到的每个特征都包含了该特征家族(例如供体、受体等)、特征类别、该特征对应的原子、特征对应序号等信息
#特征对应原子:GetAtomIds()
for i in range(0, len(feats)):
if feats[i].GetFamily() == 'Donor':
node_list = feats[i].GetAtomIds()
for i in node_list:
g.nodes[i]['donor'] = 1
elif feats[i].GetFamily() == 'Acceptor':
node_list = feats[i].GetAtomIds()
for i in node_list:
g.nodes[i]['acceptor'] = 1
# Read Edges
#读入边
#https://zhuanlan.zhihu.com/p/142935881
#下面两个循环把所有的点都连起来,做一个完全图
for i in range(0, m.GetNumAtoms()):
for j in range(0, m.GetNumAtoms()):
#获取相应的键
e_ij = m.GetBondBetweenAtoms(i, j)
#两个点有键相连
if e_ij is not None:
#添加边,边的类型,GetBondType()弄上,计算距离,coord在上面添加节点的时候放在节点的属性里面了
g.add_edge(i, j, b_type=e_ij.GetBondType(),
distance=np.linalg.norm(g.nodes[i]['coord']-g.nodes[j]['coord']))
else:
# Unbonded
g.add_edge(i, j, b_type=None,
distance=np.linalg.norm(g.nodes[i]['coord'] - g.nodes[j]['coord']))
return g , l
if __name__ == '__main__':
g1 = create_graph_grec('/home/adutta/Workspace/Datasets/Graphs/GREC/data/image1_1.gxl')
g2 = create_graph_letter('/home/adutta/Workspace/Datasets/STDGraphs/Letter/LOW/AP1_0000.gxl')
# Parse optios for downloading
parser = argparse.ArgumentParser(description='Read the specified directory, dataset and subdirectory.')
# Positional arguments
parser.add_argument('--dataset', default='GREC', nargs=1, help='Specify a dataset.')
# Optional argument
parser.add_argument('--dir', nargs=1, help='Specify the data directory.', default=['../data/'])
parser.add_argument('--subdir', nargs=1, help='Specify a subdirectory.')
args = parser.parse_args()
directory = args.dir[0]
dataset = args.dataset[0]
if dataset == 'gwhist' or dataset == 'qm9':
if args.subdir is None:
print('Error: No subdirectory mentioned for the dataset')
quit()
else:
subdir = args.subdir[0]
else:
subdir = []
print(dataset)
train_graphs, train_classes, valid_graphs, valid_classes, test_graphs, test_classes = load_dataset(directory,
dataset, subdir)
print(len(train_graphs), len(valid_graphs), len(test_graphs))
utils
#!/usr/bin/python
# -*- coding: utf-8 -*-
"""
graph_reader.py: Reads graph datasets.
Usage:
"""
import numpy as np
import networkx as nx
import random
import argparse
from rdkit import Chem
from rdkit.Chem import ChemicalFeatures
from rdkit import RDConfig
import os
from os import listdir
from os.path import isfile, join
import xml.etree.ElementTree as ET
__author__ = "Pau Riba, Anjan Dutta"
__email__ = "priba@cvc.uab.cat, adutta@cvc.uab.cat"
random.seed(2)
np.random.seed(2)
def load_dataset(directory, dataset, subdir = '01_Keypoint' ):
if dataset == 'enzymes':
file_path = join(directory, dataset)
files = [f for f in listdir(file_path) if isfile(join(file_path, f))]
classes = []
graphs = []
for i in range(len(files)):
g, c = create_graph_enzymes(join(directory, dataset, files[i]))
graphs += [g]
classes += [c]
train_graphs, train_classes, valid_graphs, valid_classes, test_graphs, test_classes = divide_datasets(graphs, classes)
elif dataset == 'mutag':
file_path = join(directory, dataset)
files = [f for f in listdir(file_path) if isfile(join(file_path, f))]
classes = []
graphs = []
for i in range(len(files)):
g, c = create_graph_mutag(join(directory, dataset, files[i]))
graphs += [g]
classes += [c]
train_graphs, train_classes, valid_graphs, valid_classes, test_graphs, test_classes = divide_datasets(graphs, classes)
elif dataset == 'MUTAG' or dataset == 'ENZYMES' or dataset == 'NCI1' or \
dataset == 'NCI109' or dataset == 'DD':
label_file = dataset + '.label'
list_file = dataset + '.list'
label_file_path = join(directory, dataset, label_file)
list_file_path = join(directory, dataset, list_file)
with open(label_file_path, 'r') as f:
l = f.read()
classes = [int(s) for s in l.split() if s.isdigit()]
with open(list_file_path, 'r') as f:
files = f.read().splitlines()
graphs = load_graphml(join(directory, dataset), files)
train_graphs, train_classes, valid_graphs, valid_classes, test_graphs, test_classes = divide_datasets(graphs, classes)
elif dataset == 'gwhist':
train_classes, train_files = read_2cols_set_files(join(directory,'Set/Train.txt'))
test_classes, test_files = read_2cols_set_files(join(directory,'Set/Test.txt'))
valid_classes, valid_files = read_2cols_set_files(join(directory,'Set/Valid.txt'))
train_classes, valid_classes, test_classes = \
create_numeric_classes(train_classes, valid_classes, test_classes)
data_dir = join(directory, 'Data/Word_Graphs/01_Skew', subdir)
train_graphs = load_gwhist(data_dir, train_files)
valid_graphs = load_gwhist(data_dir, valid_files)
test_graphs = load_gwhist(data_dir, test_files)
elif dataset == 'qm9':
file_path = join(directory, dataset, subdir)
files = [f for f in listdir(file_path) if isfile(join(file_path, f))]
data_dir = join(directory, dataset, subdir)
graphs , labels = load_qm9(data_dir, files)
# TODO: Split into train, valid and test sets and class information
idx = np.random.permutation(len(labels))
valid_graphs = [graphs[i] for i in idx[0:10000]]
valid_classes = [labels[i] for i in idx[0:10000]]
test_graphs = [graphs[i] for i in idx[10000:20000]]
test_classes = [labels[i] for i in idx[10000:20000]]
train_graphs = [graphs[i] for i in idx[20000:]]
train_classes = [labels[i] for i in idx[20000:]]
return train_graphs, train_classes, valid_graphs, valid_classes, test_graphs, test_classes
def create_numeric_classes(train_classes, valid_classes, test_classes):
classes = train_classes + valid_classes + test_classes
uniq_classes = sorted(list(set(classes)))
train_classes_ = [0] * len(train_classes)
valid_classes_ = [0] * len(valid_classes)
test_classes_ = [0] * len(test_classes)
for ix in range(len(uniq_classes)):
idx = [i for i, c in enumerate(train_classes) if c == uniq_classes[ix]]
for i in idx:
train_classes_[i] = ix
idx = [i for i, c in enumerate(valid_classes) if c == uniq_classes[ix]]
for i in idx:
valid_classes_[i] = ix
idx = [i for i, c in enumerate(test_classes) if c == uniq_classes[ix]]
for i in idx:
test_classes_[i] = ix
return train_classes_, valid_classes_, test_classes_
def load_gwhist(data_dir, files):
graphs = []
for i in range(len(files)):
g = create_graph_gwhist(join(data_dir, files[i]))
graphs += [g]
return graphs
def load_graphml(data_dir, files):
graphs = []
for i in range(len(files)):
g = nx.read_graphml(join(data_dir,files[i]))
graphs += [g]
return graphs
def load_qm9(data_dir, files):
graphs = []
labels = []
for i in range(len(files)):
g , l = xyz_graph_reader(join(data_dir, files[i]))
graphs += [g]
labels.append(l)
return graphs, labels
def read_2cols_set_files(file):
f = open(file, 'r')
lines = f.read().splitlines()
f.close()
classes = []
files = []
for line in lines:
c, f = line.split(' ')[:2]
classes += [c]
files += [f + '.gxl']
return classes, files
def read_cxl(file):
files = []
classes = []
tree_cxl = ET.parse(file)
root_cxl = tree_cxl.getroot()
for f in root_cxl.iter('print'):
files += [f.get('file')]
classes += [f.get('class')]
return classes, files
def divide_datasets(graphs, classes):
uc = list(set(classes))
tr_idx = []
va_idx = []
te_idx = []
for c in uc:
idx = [i for i, x in enumerate(classes) if x == c]
tr_idx += sorted(np.random.choice(idx, int(0.8*len(idx)), replace=False))
va_idx += sorted(np.random.choice([x for x in idx if x not in tr_idx], int(0.1*len(idx)), replace=False))
te_idx += sorted(np.random.choice([x for x in idx if x not in tr_idx and x not in va_idx], int(0.1*len(idx)), replace=False))
train_graphs = [graphs[i] for i in tr_idx]
valid_graphs = [graphs[i] for i in va_idx]
test_graphs = [graphs[i] for i in te_idx]
train_classes = [classes[i] for i in tr_idx]
valid_classes = [classes[i] for i in va_idx]
test_classes = [classes[i] for i in te_idx]
return train_graphs, train_classes, valid_graphs, valid_classes, test_graphs, test_classes
def create_graph_enzymes(file):
f = open(file, 'r')
lines = f.read().splitlines()
f.close()
# get the indices of the vertext, adj list and class
idx_vertex = lines.index("#v - vertex labels")
idx_adj_list = lines.index("#a - adjacency list")
idx_clss = lines.index("#c - Class")
# node label
vl = [int(ivl) for ivl in lines[idx_vertex+1:idx_adj_list]]
adj_list = lines[idx_adj_list+1:idx_clss]
sources = list(range(1,len(adj_list)+1))
for i in range(len(adj_list)):
if not adj_list[i]:
adj_list[i] = str(sources[i])
else:
adj_list[i] = str(sources[i])+","+adj_list[i]
g = nx.parse_adjlist(adj_list, nodetype=int, delimiter=",")
for i in range(1, g.number_of_nodes()+1):
g.nodes[i]['labels'] = np.array(vl[i-1])
c = int(lines[idx_clss+1])
return g, c
def create_graph_mutag(file):
f = open(file, 'r')
lines = f.read().splitlines()
f.close()
# get the indices of the vertext, adj list and class
idx_vertex = lines.index("#v - vertex labels")
idx_edge = lines.index("#e - edge labels")
idx_clss = lines.index("#c - Class")
# node label
vl = [int(ivl) for ivl in lines[idx_vertex+1:idx_edge]]
edge_list = lines[idx_edge+1:idx_clss]
g = nx.parse_edgelist(edge_list, nodetype=int, data=(('weight', float),), delimiter=",")
for i in range(1, g.number_of_nodes()+1):
g.nodes[i]['labels'] = np.array(vl[i-1])
c = int(lines[idx_clss+1])
return g, c
def create_graph_gwhist(file):
tree_gxl = ET.parse(file)
root_gxl = tree_gxl.getroot()
vl = []
for node in root_gxl.iter('node'):
for attr in node.iter('attr'):
if(attr.get('name') == 'x'):
x = attr.find('float').text
elif(attr.get('name') == 'y'):
y = attr.find('float').text
vl += [[x, y]]
g = nx.Graph()
for edge in root_gxl.iter('edge'):
s = edge.get('from')
s = int(s.split('_')[1])
t = edge.get('to')
t = int(t.split('_')[1])
g.add_edge(s, t)
for i in range(g.number_of_nodes()):
if i not in g.nodes:
g.add_node(i)
g.nodes[i]['labels'] = np.array(vl[i])
return g
def isfloat(value):
try:
float(value)
return True
except ValueError:
return False
def create_graph_grec(file):
tree_gxl = ET.parse(file)
root_gxl = tree_gxl.getroot()
vl = []
switch_node = {'circle': 0, 'corner': 1, 'endpoint': 2, 'intersection': 3}
switch_edge = {'arc': 0, 'arcarc': 1, 'line': 2, 'linearc': 3}
for node in root_gxl.iter('node'):
for attr in node.iter('attr'):
if (attr.get('name') == 'x'):
x = int(attr.find('Integer').text)
elif (attr.get('name') == 'y'):
y = int(attr.find('Integer').text)
elif (attr.get('name') == 'type'):
t = switch_node.get(attr.find('String').text, 4)
vl += [[x, y, t]]
g = nx.Graph()
for edge in root_gxl.iter('edge'):
s = int(edge.get('from'))
t = int(edge.get('to'))
for attr in edge.iter('attr'):
if(attr.get('name') == 'frequency'):
f = attr.find('Integer').text
elif(attr.get('name') == 'type0'):
ta = switch_edge.get(attr.find('String').text)
elif (attr.get('name') == 'angle0'):
a = attr.find('String').text
if isfloat(a):
a = float(a)
else:
a = 0.0 # TODO: The erroneous string is replaced with 0.0
g.add_edge(s, t, frequency=f, type=ta, angle=a)
for i in range(len(vl)):
if i not in g.nodes:
g.add_node(i)
g.nodes[i]['labels'] = np.array(vl[i][:3])
return g
def create_graph_letter(file):
tree_gxl = ET.parse(file)
root_gxl = tree_gxl.getroot()
vl = []
for node in root_gxl.iter('node'):
for attr in node.iter('attr'):
if (attr.get('name') == 'x'):
x = float(attr.find('float').text)
elif (attr.get('name') == 'y'):
y = float(attr.find('float').text)
vl += [[x, y]]
g = nx.Graph()
for edge in root_gxl.iter('edge'):
s = int(edge.get('from').split('_')[1])
t = int(edge.get('to').split('_')[1])
g.add_edge(s, t)
for i in range(len(vl)):
if i not in g.nodes:
g.add_node(i)
g.nodes[i]['labels'] = np.array(vl[i][:2])
return g
# Initialization of graph for QM9
def init_graph(prop):
#读取信息
prop = prop.split()
g_tag = prop[0]
g_index = int(prop[1])
g_A = float(prop[2])
g_B = float(prop[3])
g_C = float(prop[4])
#以下为labels
#论文section"4.QM9 Dataset"介绍
g_mu = float(prop[5])
g_alpha = float(prop[6])
g_homo = float(prop[7])
g_lumo = float(prop[8])
g_gap = float(prop[9])
g_r2 = float(prop[10])
g_zpve = float(prop[11])
g_U0 = float(prop[12])
g_U = float(prop[13])
g_H = float(prop[14])
g_G = float(prop[15])
g_Cv = float(prop[16])
#labels结束
labels = [g_mu, g_alpha, g_homo, g_lumo, g_gap, g_r2, g_zpve, g_U0, g_U, g_H, g_G, g_Cv]
#返回networkx类型的图,标签list
return nx.Graph(tag=g_tag, index=g_index, A=g_A, B=g_B, C=g_C, mu=g_mu, alpha=g_alpha, homo=g_homo,
lumo=g_lumo, gap=g_gap, r2=g_r2, zpve=g_zpve, U0=g_U0, U=g_U, H=g_H, G=g_G, Cv=g_Cv), labels
# XYZ file reader for QM9 dataset
def xyz_graph_reader(graph_file):
#数据例子:
# https://www.kaggle.com/zaharch/quantum-machine-9-aka-qm9?select=dsgdb9nsd_000001.xyz
'''
第一行 5
第二行 gdb 1 157.7118 157.70997 157.70699 0. 13.21 -0.3877 0.1171 0.5048 35.3641 0.044749 -40.47893 -40.476062 -40.475117 -40.498597 6.469
第三行 C -0.0126981359 1.0858041578 0.0080009958 -0.535689
第四行 H 0.002150416 -0.0060313176 0.0019761204 0.133921
第五行 H 1.0117308433 1.4637511618 0.0002765748 0.133922
第六行 H -0.540815069 1.4475266138 -0.8766437152 0.133923
第七行 H -0.5238136345 1.4379326443 0.9063972942 0.133923
第八行 1341.307 1341.3284 1341.365 1562.6731 1562.7453 3038.3205 3151.6034 3151.6788 3151.7078
第九行 C C
第十行 InChI=1S/CH4/h1H4 InChI=1S/CH4/h1H4
'''
with open(graph_file,'r') as f:
# Number of atoms
# 第一行原子数量,上例中是5
na = int(f.readline())
# Graph properties
#第二行,图的性质(ground truth labels)
properties = f.readline()
#得到networkx类型的图,内含标签list
g, l = init_graph(properties)
atom_properties = []
# Atoms properties
#根据第一行读到的原子数量na,读取na行得到每个原子的信息
for i in range(na):
a_properties = f.readline()#每一行都存为一个list,
a_properties = a_properties.replace('.*^', 'e')
a_properties = a_properties.replace('*^', 'e')
a_properties = a_properties.split()
atom_properties.append(a_properties)#atom_properties是list套list
# Frequencies
#这一行信息互略
f.readline()
# SMILES(Simplified molecular input line entry specification),简化分子线性输入规范,是一种用ASCII字符串明确描述分子结构的规范。
smiles = f.readline()
smiles = smiles.split()
smiles = smiles[0]
#参考rdkit包
# https://www.rdkit.org/docs/source/rdkit.Chem.html
#smiles转换为分子对象
# https://zhuanlan.zhihu.com/p/82497166
m = Chem.MolFromSmiles(smiles)
m = Chem.AddHs(m)#按原文加入氢原子
#from rdkit import RDConfig
#https://zhuanlan.zhihu.com/p/141908982
#导入特征库
fdef_name = os.path.join(RDConfig.RDDataDir, 'BaseFeatures.fdef')
#创建特征工厂
factory = ChemicalFeatures.BuildFeatureFactory(fdef_name)
#计算化学特征
feats = factory.GetFeaturesForMol(m)
# Create nodes
#读入点
for i in range(0, m.GetNumAtoms()):
# https://zhuanlan.zhihu.com/p/143111689
# 通过索引获取原子
atom_i = m.GetAtomWithIdx(i)
#获取原子符号:Getsymbol()
#获取原子序号:GetAtomicNum()
#获取原子杂化方式:GetHybridization()
# atom properties是上面处理的原子的信息
g.add_node(i, a_type=atom_i.GetSymbol(), a_num=atom_i.GetAtomicNum(), acceptor=0, donor=0,
aromatic=atom_i.GetIsAromatic(), hybridization=atom_i.GetHybridization(),
num_h=atom_i.GetTotalNumHs(), coord=np.array(atom_properties[i][1:4]).astype(np.float),
pc=float(atom_properties[i][4]))
# https://zhuanlan.zhihu.com/p/141908982
#遍历化学特征feats,找到包含供体、受体的原子的id(GetAtomIds()),把这些原子id放到node_list,然后遍历node_list,分别加donor和acceptor属性
#搜索到的每个特征都包含了该特征家族(例如供体、受体等)、特征类别、该特征对应的原子、特征对应序号等信息
#特征对应原子:GetAtomIds()
for i in range(0, len(feats)):
if feats[i].GetFamily() == 'Donor':
node_list = feats[i].GetAtomIds()
for i in node_list:
g.nodes[i]['donor'] = 1
elif feats[i].GetFamily() == 'Acceptor':
node_list = feats[i].GetAtomIds()
for i in node_list:
g.nodes[i]['acceptor'] = 1
# Read Edges
#读入边
#https://zhuanlan.zhihu.com/p/142935881
#下面两个循环把所有的点都连起来,做一个完全图
for i in range(0, m.GetNumAtoms()):
for j in range(0, m.GetNumAtoms()):
#获取相应的键
e_ij = m.GetBondBetweenAtoms(i, j)
#两个点有键相连
if e_ij is not None:
#添加边,边的类型,GetBondType()弄上,计算距离,coord在上面添加节点的时候放在节点的属性里面了
g.add_edge(i, j, b_type=e_ij.GetBondType(),
distance=np.linalg.norm(g.nodes[i]['coord']-g.nodes[j]['coord']))
else:
# Unbonded
g.add_edge(i, j, b_type=None,
distance=np.linalg.norm(g.nodes[i]['coord'] - g.nodes[j]['coord']))
return g , l
if __name__ == '__main__':
g1 = create_graph_grec('/home/adutta/Workspace/Datasets/Graphs/GREC/data/image1_1.gxl')
g2 = create_graph_letter('/home/adutta/Workspace/Datasets/STDGraphs/Letter/LOW/AP1_0000.gxl')
# Parse optios for downloading
parser = argparse.ArgumentParser(description='Read the specified directory, dataset and subdirectory.')
# Positional arguments
parser.add_argument('--dataset', default='GREC', nargs=1, help='Specify a dataset.')
# Optional argument
parser.add_argument('--dir', nargs=1, help='Specify the data directory.', default=['../data/'])
parser.add_argument('--subdir', nargs=1, help='Specify a subdirectory.')
args = parser.parse_args()
directory = args.dir[0]
dataset = args.dataset[0]
if dataset == 'gwhist' or dataset == 'qm9':
if args.subdir is None:
print('Error: No subdirectory mentioned for the dataset')
quit()
else:
subdir = args.subdir[0]
else:
subdir = []
print(dataset)
train_graphs, train_classes, valid_graphs, valid_classes, test_graphs, test_classes = load_dataset(directory,
dataset, subdir)
print(len(train_graphs), len(valid_graphs), len(test_graphs))
MPNN
#!/usr/bin/python
# -*- coding: utf-8 -*-
from MessageFunction import MessageFunction
from UpdateFunction import UpdateFunction
from ReadoutFunction import ReadoutFunction
import torch
import torch.nn as nn
from torch.autograd import Variable
__author__ = "Pau Riba, Anjan Dutta"
__email__ = "priba@cvc.uab.cat, adutta@cvc.uab.cat"
class MPNN(nn.Module):
"""
MPNN as proposed by Gilmer et al..
This class implements the whole Gilmer et al. model following the functions Message, Update and Readout.
Parameters
----------
in_n : int list
Sizes for the node and edge features.
hidden_state_size : int
Size of the hidden states (the input will be padded with 0's to this size).
message_size : int
Message function output vector size.
n_layers : int
Number of iterations Message+Update (weight tying).
l_target : int
Size of the output.
type : str (Optional)
Classification | [Regression (default)]. If classification, LogSoftmax layer is applied to the output vector.
"""
def __init__(self, in_n, hidden_state_size, message_size, n_layers, l_target, type='regression'):
super(MPNN, self).__init__()
# Define message
#in_n:点的特征维度,边的特征维度
#hidden_state_size.73
#message_size.73
self.m = nn.ModuleList(
[MessageFunction('mpnn', args={'edge_feat': in_n[1], 'in': hidden_state_size, 'out': message_size})])
# Define Update
self.u = nn.ModuleList([UpdateFunction('mpnn',
args={'in_m': message_size,
'out': hidden_state_size})])
# Define Readout
self.r = ReadoutFunction('mpnn',
args={'in': hidden_state_size,#73
'target': l_target})#这里是13分类,所以是13
#回归任务
self.type = type
self.args = {}
self.args['out'] = hidden_state_size
self.n_layers = n_layers#3
def forward(self, g, h_in, e):
h = []#列表,保存点的embedding,h0表示输入层点的embedding,h1表示第一层点的embedding,ht表示第T层点的embedding
# Padding to some larger dimension d
h_t = torch.cat([h_in, Variable(
torch.zeros(h_in.size(0), h_in.size(1), self.args['out'] - h_in.size(2)).type_as(h_in.data))], 2)
h.append(h_t.clone())
# Layer
# 每一层进行循环
for t in range(0, self.n_layers):
#utils.py中的collate_g函数已经按batch放好了:
#g,h,e,target
#g:[bs,N,N]
#h:[bs,N,d_v]
#e:[bs,N,N,d_e]
#e_aux:[bs*N*N,d_e]
e_aux = e.view(-1, e.size(3))
#h_aux:[bs *N,d_v]
h_aux = h[t].view(-1, h[t].size(2))
#1. Message Function里面的forward
#m:[ bs*N*N,d_v]
m = self.m[0].forward(h[t], h_aux, e_aux)
#m:[bs,N,N,d_v]
m = m.view(h[0].size(0), h[0].size(1), -1, m.size(1))
# Nodes without edge set message to 0
#m:[bs,N,N,d_v]
m = torch.unsqueeze(g, 3).expand_as(m) * m
#sum([bs,N,N,d_v],1)后squeeze第一个N就没有了
#m:[bs,N,d_v]
m = torch.squeeze(torch.sum(m, 1))
#2. Update Function
#h_t:[ bs,N,d_v]
h_t = self.u[0].forward(h[t], m)
# Delete virtual nodes
# h_t = (torch.sum(h_in, 2).expand_as(h_t) > 0).type_as(h_t) * h_t
h_t = (torch.sum(h_in, 2)[..., None].expand_as(h_t) > 0).type_as(h_t) * h_t
h.append(h_t)
# 3.Readout
res = self.r.forward(h)
if self.type == 'classification':
res = nn.LogSoftmax()(res)
return res
MessageFunction
#!/usr/bin/python
# -*- coding: utf-8 -*-
"""
MessageFunction.py: Propagates a message depending on two nodes and their common edge.
Usage:
"""
from __future__ import print_function
# Own modules
import datasets
from models.nnet import NNet
import numpy as np
import os
import argparse
import time
import torch
import torch.nn as nn
from torch.autograd.variable import Variable
__author__ = "Pau Riba, Anjan Dutta"
__email__ = "priba@cvc.uab.cat, adutta@cvc.uab.cat"
class MessageFunction(nn.Module):
# Constructor
def __init__(self, message_def='duvenaud', args={}):
super(MessageFunction, self).__init__()
#指定m duvenaud,m ggnn,m mpnn哪种类型
self.m_definition = ''
self.m_function = None
self.args = {}
#指定类型和参数,message_def这里用的是mpnn
self.__set_message(message_def, args)
# Message from h_v to h_w through e_vw
def forward(self, h_v, h_w, e_vw, args=None):
return self.m_function(h_v, h_w, e_vw, args)
# Set a message function
def __set_message(self, message_def, args={}):
self.m_definition = message_def.lower()#大小写转换
#指定m_function使用哪个函数
self.m_function = {
'duvenaud': self.m_duvenaud,
'ggnn': self.m_ggnn,
'intnet': self.m_intnet,
'mpnn': self.m_mpnn,
'mgc': self.m_mgc,
'bruna': self.m_bruna,
'defferrard': self.m_deff,
'kipf': self.m_kipf
}.get(self.m_definition, None)
if self.m_function is None:
print('WARNING!: Message Function has not been set correctly\n\tIncorrect definition ' + message_def)
quit()
#指定参数初始化方法
init_parameters = {
'duvenaud': self.init_duvenaud,
'ggnn': self.init_ggnn,
'intnet': self.init_intnet,
'mpnn': self.init_mpnn
}.get(self.m_definition, lambda x: (nn.ParameterList([]), nn.ModuleList([]), {}))
#初始化参数
self.learn_args, self.learn_modules, self.args = init_parameters(args)
#输出的m_i向量维度
self.m_size = {
'duvenaud': self.out_duvenaud,
'ggnn': self.out_ggnn,
'intnet': self.out_intnet,
'mpnn': self.out_mpnn
}.get(self.m_definition, None)
# Get the name of the used message function
def get_definition(self):
return self.m_definition
# Get the message function arguments
def get_args(self):
return self.args
# Get Output size
def get_out_size(self, size_h, size_e, args=None):
return self.m_size(size_h, size_e, args)
# Definition of various state of the art message functions
# Duvenaud et al. (2015), Convolutional Networks for Learning Molecular Fingerprints
def m_duvenaud(self, h_v, h_w, e_vw, args):
m = torch.cat([h_w, e_vw], 2)
return m
def out_duvenaud(self, size_h, size_e, args):
return size_h + size_e
def init_duvenaud(self, params):
learn_args = []
learn_modules = []
args = {}
return nn.ParameterList(learn_args), nn.ModuleList(learn_modules), args
# Li et al. (2016), Gated Graph Neural Networks (GG-NN)
def m_ggnn(self, h_v, h_w, e_vw, opt={}):
m = Variable(torch.zeros(h_w.size(0), h_w.size(1), self.args['out']).type_as(h_w.data))
for w in range(h_w.size(1)):
if torch.nonzero(e_vw[:, w, :].data).size():
for i, el in enumerate(self.args['e_label']):
ind = (el == e_vw[:,w,:]).type_as(self.learn_args[0][i])
parameter_mat = self.learn_args[0][i][None, ...].expand(h_w.size(0), self.learn_args[0][i].size(0),
self.learn_args[0][i].size(1))
m_w = torch.transpose(torch.bmm(torch.transpose(parameter_mat, 1, 2),
torch.transpose(torch.unsqueeze(h_w[:, w, :], 1),
1, 2)), 1, 2)
m_w = torch.squeeze(m_w)
m[:,w,:] = ind.expand_as(m_w)*m_w
return m
def out_ggnn(self, size_h, size_e, args):
return self.args['out']
def init_ggnn(self, params):
learn_args = []
learn_modules = []
args = {}
args['e_label'] = params['e_label']
args['in'] = params['in']
args['out'] = params['out']
# Define a parameter matrix A for each edge label.
learn_args.append(nn.Parameter(torch.randn(len(params['e_label']), params['in'], params['out'])))
return nn.ParameterList(learn_args), nn.ModuleList(learn_modules), args
# Battaglia et al. (2016), Interaction Networks
def m_intnet(self, h_v, h_w, e_vw, args):
m = torch.cat([h_v[:, None, :].expand_as(h_w), h_w, e_vw], 2)
b_size = m.size()
m = m.view(-1, b_size[2])
m = self.learn_modules[0](m)
m = m.view(b_size[0], b_size[1], -1)
return m
def out_intnet(self, size_h, size_e, args):
return self.args['out']
def init_intnet(self, params):
learn_args = []
learn_modules = []
args = {}
args['in'] = params['in']
args['out'] = params['out']
learn_modules.append(NNet(n_in=params['in'], n_out=params['out']))
return nn.ParameterList(learn_args), nn.ModuleList(learn_modules), args
# Gilmer et al. (2017), Neural Message Passing for Quantum Chemistry
def m_mpnn(self, h_v, h_w, e_vw, opt={}):
# Matrices for each edge
#h_v:[bs,N,d_v]
#h_w:[bs*N,d_v]
#e_vw:[bs*N*N,d_e]
#edge output:[bs*N*N,d_v*d_v]
#learn_modules就是论文5.1节,edge network的矩阵A,是要学习的参数
#如果把e_vw看做输入,A看做NN的参数,这个NN的输入输出是:(看init_mpnn)
#(n_in=params['edge_feat'], n_out=(params['in']*params['out'])
#也就是输入边的特征维度d_e,输出是73*73
#e_vw:[bs*N*N,d_e]经过这个NN后维度就变成了[bs*N*N,d_v*d_v]也就是[bs*N*N,73*73]
edge_output = self.learn_modules[0](e_vw)
#把维度拆开一下
#edge output:[bs*N*N,d_v,d_v]
edge_output = edge_output.view(-1, self.args['out'], self.args['in'])
# tensor维度中使用None
# https://blog.csdn.net/jmu201521121021/article/details/103773501
#None是在最后一维扩展一个维度,使得h_w原来的2维变成3维,这三个维度分别为:
#h_w.size(0):bs*N
# h_v.size(1):N
# h_v.size(1)):d_v
#所以h_w_rows维度是:[bs*N,N,d_v]
h_w_rows = h_w[..., None].expand(h_w.size(0), h_w.size(1), h_v.size(1)).contiguous()
#把前面两维合起来:
#h_w_rows维度是:[bs*N*N,d_v]
h_w_rows = h_w_rows.view(-1, self.args['in'])
#h_w_rows为了要和output相乘,这里要unsqueeze后加1维,变成:[bs*N*N,d_v,1]
#BMM时bs*N*N这个维度是不动的[d_v,d_v].[d_v,1]的结果是[d_v,1]
#最后bmm结果h_multiply:[bs*N*N,d_v,1]
h_multiply = torch.bmm(edge_output, torch.unsqueeze(h_w_rows,2))
#m_new:[bs*N*N,d_v]
m_new = torch.squeeze(h_multiply)
return m_new
def out_mpnn(self, size_h, size_e, args):
return self.args['out']
def init_mpnn(self, params):
learn_args = []
learn_modules = []
args = {}
#params={'edge_feat': in_n[1], 'in': hidden_state_size, 'out': message_size}
args['in'] = params['in']#73
args['out'] = params['out']#73
# Define a parameter matrix A for each edge label.
#论文5.1节,edge network
#定义矩阵A
learn_modules.append(NNet(n_in=params['edge_feat'], n_out=(params['in']*params['out'])))
return nn.ParameterList(learn_args), nn.ModuleList(learn_modules), args
# Kearnes et al. (2016), Molecular Graph Convolutions
def m_mgc(self, h_v, h_w, e_vw, args):
m = e_vw
return m
# Laplacian based methods
# Bruna et al. (2013)
def m_bruna(self, h_v, h_w, e_vw, args):
# TODO
m = []
return m
# Defferrard et al. (2016)
def m_deff(self, h_v, h_w, e_vw, args):
# TODO
m = []
return m
# Kipf & Welling (2016)
def m_kipf(self, h_v, h_w, e_vw, args):
# TODO
m = []
return m
if __name__ == '__main__':
# Parse optios for downloading
parser = argparse.ArgumentParser(description='QM9 Object.')
# Optional argument
parser.add_argument('--root', nargs=1, help='Specify the data directory.', default=['./data/qm9/dsgdb9nsd/'])
args = parser.parse_args()
root = args.root[0]
files = [f for f in os.listdir(root) if os.path.isfile(os.path.join(root, f))]
idx = np.random.permutation(len(files))
idx = idx.tolist()
valid_ids = [files[i] for i in idx[0:10000]]
test_ids = [files[i] for i in idx[10000:20000]]
train_ids = [files[i] for i in idx[20000:]]
data_train = datasets.Qm9(root, train_ids)
data_valid = datasets.Qm9(root, valid_ids)
data_test = datasets.Qm9(root, test_ids)
# Define message
m = MessageFunction('duvenaud')
print(m.get_definition())
start = time.time()
# Select one graph
g_tuple, l = data_train[0]
g, h_t, e = g_tuple
m_t = {}
for v in g.nodes_iter():
neigh = g.neighbors(v)
m_neigh = type(h_t)
for w in neigh:
if (v,w) in e:
e_vw = e[(v, w)]
else:
e_vw = e[(w, v)]
m_v = m.forward(h_t[v], h_t[w], e_vw)
if len(m_neigh):
m_neigh += m_v
else:
m_neigh = m_v
m_t[v] = m_neigh
end = time.time()
print('Input nodes')
print(h_t)
print('Message')
print(m_t)
print('Time')
print(end - start)
UpdateFunction
#!/usr/bin/python
# -*- coding: utf-8 -*-
"""
UpdateFunction.py: Updates the nodes using the previous state and the message.
Usage:
"""
from __future__ import print_function
# Own modules
import datasets
from MessageFunction import MessageFunction
from models.nnet import NNet
import numpy as np
import time
import os
import argparse
import torch
import torch.nn as nn
import torch.nn.functional as F
from torch.autograd.variable import Variable
#dtype = torch.cuda.FloatTensor
dtype = torch.FloatTensor
__author__ = "Pau Riba, Anjan Dutta"
__email__ = "priba@cvc.uab.cat, adutta@cvc.uab.cat"
class UpdateFunction(nn.Module):
# Constructor
def __init__(self, update_def='nn', args={}):
super(UpdateFunction, self).__init__()
#指定u_duvenaud,u_ggnn,u_mpnn哪种类型
self.u_definition = ''
self.u_function = None
self.args = {}
self.__set_update(update_def, args)
# Update node hv given message mv
def forward(self, h_v, m_v, opt={}):
return self.u_function(h_v, m_v, opt)
# Set update function
def __set_update(self, update_def, args):
self.u_definition = update_def.lower()
#指定u_function使用哪个函数
self.u_function = {
'duvenaud': self.u_duvenaud,
'ggnn': self.u_ggnn,
'intnet': self.u_intnet,
'mpnn': self.u_mpnn
}.get(self.u_definition, None)
if self.u_function is None:
print('WARNING!: Update Function has not been set correctly\n\tIncorrect definition ' + update_def)
init_parameters = {
'duvenaud': self.init_duvenaud,
'ggnn': self.init_ggnn,
'intnet': self.init_intnet,
'mpnn': self.init_mpnn
}.get(self.u_definition, lambda x: (nn.ParameterList([]), nn.ModuleList([]), {}))
self.learn_args, self.learn_modules, self.args = init_parameters(args)
# Get the name of the used update function
def get_definition(self):
return self.u_definition
# Get the update function arguments
def get_args(self):
return self.args
## Definition of various state of the art update functions
# Duvenaud
def u_duvenaud(self, h_v, m_v, opt):
param_sz = self.learn_args[0][opt['deg']].size()
parameter_mat = torch.t(self.learn_args[0][opt['deg']])[None, ...].expand(m_v.size(0), param_sz[1], param_sz[0])
aux = torch.bmm(parameter_mat, torch.transpose(m_v, 1, 2))
return torch.transpose(torch.nn.Sigmoid()(aux), 1, 2)
def init_duvenaud(self, params):
learn_args = []
learn_modules = []
args = {}
# Filter degree 0 (the message will be 0 and therefore there is no update
args['deg'] = [i for i in params['deg'] if i!=0]
args['in'] = params['in']
args['out'] = params['out']
# Define a parameter matrix H for each degree.
learn_args.append(torch.nn.Parameter(torch.randn(len(args['deg']), args['in'], args['out'])))
return nn.ParameterList(learn_args), nn.ModuleList(learn_modules), args
# GG-NN, Li et al.
def u_ggnn(self, h_v, m_v, opt={}):
h_v.contiguous()
m_v.contiguous()
h_new = self.learn_modules[0](torch.transpose(m_v, 0, 1), torch.unsqueeze(h_v, 0))[0] # 0 or 1???
return torch.transpose(h_new, 0, 1)
def init_ggnn(self, params):
learn_args = []
learn_modules = []
args = {}
args['in_m'] = params['in_m']
args['out'] = params['out']
# GRU
learn_modules.append(nn.GRU(params['in_m'], params['out']))
return nn.ParameterList(learn_args), nn.ModuleList(learn_modules), args
# Battaglia et al. (2016), Interaction Networks
def u_intnet(self, h_v, m_v, opt):
if opt['x_v'].ndimension():
input_tensor = torch.cat([h_v, opt['x_v'], torch.squeeze(m_v)], 1)
else:
input_tensor = torch.cat([h_v, torch.squeeze(m_v)], 1)
return self.learn_modules[0](input_tensor)
def init_intnet(self, params):
learn_args = []
learn_modules = []
args = {}
args['in'] = params['in']
args['out'] = params['out']
learn_modules.append(NNet(n_in=params['in'], n_out=params['out']))
return nn.ParameterList(learn_args), nn.ModuleList(learn_modules), args
def u_mpnn(self, h_v, m_v, opt={}):
#h_v:[bs,N,d_v]
#m_v:[bs,N,d_v]
#h_in:[bs*N,d_v]
h_in = h_v.view(-1,h_v.size(2))
#m_in:[bs*N,d_v]
m_in = m_v.view(-1,m_v.size(2))
# https://pytorch.org/docs/stable/generated/torch.nn.GRU.html
# https://www.jianshu.com/p/b942e65cb0a3
#m_in as GRU input x_t,h_in as GRU hidden state h_t
#GRU输入大小是三维tensor(seq_len,batch_size,input_dim]
#输入序列seq len=1
#h_new=[1,bs*N,d_v]
h_new = self.learn_modules[0](m_in[None,...],h_in[None,...])[0] # 0 or 1???0是输出GRU每一个时间步的output,1代表GRU的每个时间步的隐藏层,GGNN用的隐藏层来表示点的embedding,应该用1
#把h_new的第一维去掉后把第二维拆开:[bs,N,d_v]
return torch.squeeze(h_new).view(h_v.size())
def init_mpnn(self, params):
learn_args = []
learn_modules = []
args = {}
args['in_m'] = params['in_m']#73
args['out'] = params['out']#73
# GRU
# 使用GRU作为update函数,'in_m'做为GRU的x_dim,'out'作为GRU的隐层h_dim(,layer_num)
learn_modules.append(nn.GRU(params['in_m'], params['out']))
return nn.ParameterList(learn_args), nn.ModuleList(learn_modules), args
if __name__ == '__main__':
# Parse optios for downloading
parser = argparse.ArgumentParser(description='QM9 Object.')
# Optional argument
parser.add_argument('--root', nargs=1, help='Specify the data directory.', default=['./data/qm9/dsgdb9nsd/'])
args = parser.parse_args()
root = args.root[0]
files = [f for f in os.listdir(root) if os.path.isfile(os.path.join(root, f))]
idx = np.random.permutation(len(files))
idx = idx.tolist()
valid_ids = [files[i] for i in idx[0:10000]]
test_ids = [files[i] for i in idx[10000:20000]]
train_ids = [files[i] for i in idx[20000:]]
data_train = datasets.Qm9(root, train_ids)
data_valid = datasets.Qm9(root, valid_ids)
data_test = datasets.Qm9(root, test_ids)
print('STATS')
# d = datasets.utils.get_graph_stats(data_test, 'degrees')
d = [1, 2, 3, 4]
print('Message')
## Define message
m = MessageFunction('duvenaud')
## Parameters for the update function
# Select one graph
g_tuple, l = data_train[0]
g, h_t, e = g_tuple
m_v = m.forward(h_t[0], h_t[1], e[list(e.keys())[0]])
in_n = len(m_v)
out_n = 30
print('Update')
## Define Update
u = UpdateFunction('duvenaud', args={'deg': d, 'in': in_n , 'out': out_n})
print(m.get_definition())
print(u.get_definition())
start = time.time()
# Select one graph
g_tuple, l = data_train[0]
g, h_t, e = g_tuple
h_t1 = {}
for v in g.nodes_iter():
neigh = g.neighbors(v)
m_neigh = dtype()
for w in neigh:
if (v, w) in e:
e_vw = e[(v, w)]
else:
e_vw = e[(w, v)]
m_v = m.forward(h_t[v], h_t[w], e_vw)
if len(m_neigh):
m_neigh += m_v
else:
m_neigh = m_v
# Duvenaud
opt = {'deg': len(neigh)}
h_t1[v] = u.forward(h_t[v], m_neigh, opt)
end = time.time()
print('Input nodes')
print(h_t)
print('Message')
print(h_t1)
print('Time')
print(end - start)
ReadoutFunction
#!/usr/bin/python
# -*- coding: utf-8 -*-
"""
MessageFunction.py: Propagates a message depending on two nodes and their common edge.
Usage:
"""
from __future__ import print_function
# Own modules
import datasets
from MessageFunction import MessageFunction
from UpdateFunction import UpdateFunction
from models.nnet import NNet
import time
import torch
import torch.nn as nn
import os
import argparse
import numpy as np
from torch.autograd.variable import Variable
#dtype = torch.cuda.FloatTensor
dtype = torch.FloatTensor
__author__ = "Pau Riba, Anjan Dutta"
__email__ = "priba@cvc.uab.cat, adutta@cvc.uab.cat"
class ReadoutFunction(nn.Module):
# Constructor
def __init__(self, readout_def='nn', args={}):
super(ReadoutFunction, self).__init__()
#指定rduvenaud,r ggnn,r mpnn种类型
self.r_definition = ''
self.r_function = None
self.args = {}
self.__set_readout(readout_def, args)
# Readout graph given node values at las layer
def forward(self, h_v):
return self.r_function(h_v)
# Set a readout function
def __set_readout(self, readout_def, args):
self.r_definition = readout_def.lower()
#指定r_function使用哪个函数
self.r_function = {
'duvenaud': self.r_duvenaud,
'ggnn': self.r_ggnn,
'intnet': self.r_intnet,
'mpnn': self.r_mpnn
}.get(self.r_definition, None)
if self.r_function is None:
print('WARNING!: Readout Function has not been set correctly\n\tIncorrect definition ' + readout_def)
quit()
#指定参数初始化方法
init_parameters = {
'duvenaud': self.init_duvenaud,
'ggnn': self.init_ggnn,
'intnet': self.init_intnet,
'mpnn': self.init_mpnn
}.get(self.r_definition, lambda x: (nn.ParameterList([]), nn.ModuleList([]), {}))
#初始化参数
self.learn_args, self.learn_modules, self.args = init_parameters(args)
# Get the name of the used readout function
def get_definition(self):
return self.r_definition
## Definition of various state of the art update functions
# Duvenaud
def r_duvenaud(self, h):
# layers
aux = []
for l in range(len(h)):
param_sz = self.learn_args[l].size()
parameter_mat = torch.t(self.learn_args[l])[None, ...].expand(h[l].size(0), param_sz[1],
param_sz[0])
aux.append(torch.transpose(torch.bmm(parameter_mat, torch.transpose(h[l], 1, 2)), 1, 2))
for j in range(0, aux[l].size(1)):
# Mask whole 0 vectors
aux[l][:, j, :] = nn.Softmax()(aux[l][:, j, :].clone())*(torch.sum(aux[l][:, j, :] != 0, 1) > 0).expand_as(aux[l][:, j, :]).type_as(aux[l])
aux = torch.sum(torch.sum(torch.stack(aux, 3), 3), 1)
return self.learn_modules[0](torch.squeeze(aux))
def init_duvenaud(self, params):
learn_args = []
learn_modules = []
args = {}
args['out'] = params['out']
# Define a parameter matrix W for each layer.
for l in range(params['layers']):
learn_args.append(nn.Parameter(torch.randn(params['in'][l], params['out'])))
# learn_modules.append(nn.Linear(params['out'], params['target']))
learn_modules.append(NNet(n_in=params['out'], n_out=params['target']))
return nn.ParameterList(learn_args), nn.ModuleList(learn_modules), args
# GG-NN, Li et al.
def r_ggnn(self, h):
aux = Variable( torch.Tensor(h[0].size(0), self.args['out']).type_as(h[0].data).zero_() )
# For each graph
for i in range(h[0].size(0)):
nn_res = nn.Sigmoid()(self.learn_modules[0](torch.cat([h[0][i,:,:], h[-1][i,:,:]], 1)))*self.learn_modules[1](h[-1][i,:,:])
# Delete virtual nodes
nn_res = (torch.sum(h[0][i,:,:],1).expand_as(nn_res)>0).type_as(nn_res)* nn_res
aux[i,:] = torch.sum(nn_res,0)
return aux
def init_ggnn(self, params):
learn_args = []
learn_modules = []
args = {}
# i
learn_modules.append(NNet(n_in=2*params['in'], n_out=params['target']))
# j
learn_modules.append(NNet(n_in=params['in'], n_out=params['target']))
args['out'] = params['target']
return nn.ParameterList(learn_args), nn.ModuleList(learn_modules), args
# Battaglia et al. (2016), Interaction Networks
def r_intnet(self, h):
aux = torch.sum(h[-1],1)
return self.learn_modules[0](aux)
def init_intnet(self, params):
learn_args = []
learn_modules = []
args = {}
learn_modules.append(NNet(n_in=params['in'], n_out=params['target']))
return nn.ParameterList(learn_args), nn.ModuleList(learn_modules), args
def r_mpnn(self, h):
#h:list of[bs,N,d_v]
#h[0].size(0):bs
#self.args['out']:1_target=labels数量
aux = Variable( torch.Tensor(h[0].size(0), self.args['out']).type_as(h[0].data).zero_() )
# For each graph
for i in range(h[0].size(0)):
#论文公式(4),h[0]是h_v^0,h[-1]是h_v^T
nn_res = nn.Sigmoid()(self.learn_modules[0](torch.cat([h[0][i,:,:], h[-1][i,:,:]], 1)))*self.learn_modules[1](h[-1][i,:,:])
# Delete virtual nodes
# nn_res = (torch.sum(h[0][i,:,:],1).expand_as(nn_res)>0).type_as(nn_res)* nn_res
nn_res = (torch.sum(h[0][i,:,:],1)[...,None].expand_as(nn_res)>0).type_as(nn_res)* nn_res
aux[i,:] = torch.sum(nn_res,0)
#[bs,1_target]
return aux
def init_mpnn(self, params):
learn_args = []
learn_modules = []
args = {}
# i
#采用GGNN中的readout函数,论文中的公式(4)
#i:(h_v^T,h_v^0)拼接,T步后的点embedding和初始的点embedding做拼接,所以这里要乘2
learn_modules.append(NNet(n_in=2*params['in'], n_out=params['target']))
# j:(h_v^T)
learn_modules.append(NNet(n_in=params['in'], n_out=params['target']))
#最后的graph embedding R为i,j的element-wise multiplications
# ground truth labels
args['out'] = params['target']
return nn.ParameterList(learn_args), nn.ModuleList(learn_modules), args
if __name__ == '__main__':
# Parse optios for downloading
parser = argparse.ArgumentParser(description='QM9 Object.')
# Optional argument
parser.add_argument('--root', nargs=1, help='Specify the data directory.', default=['./data/qm9/dsgdb9nsd/'])
args = parser.parse_args()
root = args.root[0]
files = [f for f in os.listdir(root) if os.path.isfile(os.path.join(root, f))]
idx = np.random.permutation(len(files))
idx = idx.tolist()
valid_ids = [files[i] for i in idx[0:10000]]
test_ids = [files[i] for i in idx[10000:20000]]
train_ids = [files[i] for i in idx[20000:]]
data_train = datasets.Qm9(root, train_ids)
data_valid = datasets.Qm9(root, valid_ids)
data_test = datasets.Qm9(root, test_ids)
# d = datasets.utils.get_graph_stats(data_train, 'degrees')
d = [1, 2, 3, 4]
## Define message
m = MessageFunction('duvenaud')
## Parameters for the update function
# Select one graph
g_tuple, l = data_train[0]
g, h_t, e = g_tuple
m_v = m.forward(h_t[0], h_t[1], e[list(e.keys())[0]])
in_n = len(m_v)
out_n = 30
## Define Update
u = UpdateFunction('duvenaud', args={'deg': d, 'in': in_n, 'out': out_n})
in_n = len(h_t[0])
## Define Readout
r = ReadoutFunction('duvenaud', args={'layers': 2, 'in': [in_n, out_n], 'out': 50, 'target': len(l)})
print(m.get_definition())
print(u.get_definition())
print(r.get_definition())
start = time.time()
# Layers
h = []
# Select one graph
g_tuple, l = data_train[0]
g, h_in, e = g_tuple
h.append(h_in)
# Layer
t = 1
h.append({})
for v in g.nodes_iter():
neigh = g.neighbors(v)
m_neigh = dtype()
for w in neigh:
if (v, w) in e:
e_vw = e[(v, w)]
else:
e_vw = e[(w, v)]
m_v = m.forward(h[t-1][v], h[t-1][w], e_vw)
if len(m_neigh):
m_neigh += m_v
else:
m_neigh = m_v
# Duvenaud
opt = {'deg': len(neigh)}
h[t][v] = u.forward(h[t-1][v], m_neigh, opt)
# Readout
res = r.forward(h)
end = time.time()
print(res)
print('Time')
print(end - start)




























 429
429

 被折叠的 条评论
为什么被折叠?
被折叠的 条评论
为什么被折叠?










