UPGMA (Unweighted Pair Group Method with Arithmetic Mean) is a simple agglomerative (bottom-up) hierarchical clustering method. The method is generally attributed to Sokal and Michener.[1]
The UPGMA method is similar to its weighted variant, the WPGMA method.
Contents
Algorithm
The UPGMA algorithm constructs a rooted tree (dendrogram) that reflects the structure present in a pairwise similarity matrix (or a dissimilarity matrix). At each step, the nearest two clusters are combined into a higher-level cluster. The distance between any two clusters A and B, each of size (i.e., cardinality) 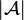 and
and 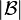 , is taken to be the average of all distances
, is taken to be the average of all distances  between pairs of objects
between pairs of objects 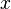 in
in  and
and 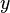 in
in 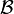 , that is, the mean distance between elements of each cluster:
, that is, the mean distance between elements of each cluster:
In other words, at each clustering step, the updated distance between the joined clusters  and a new cluster
and a new cluster 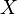 is given by the proportional averaging of the
is given by the proportional averaging of the  and
and  distances:
distances:

The UPGMA algorithm produces rooted dendrograms and requires a constant-rate assumption - that is, it assumes an ultrametric tree in which the distances from the root to every branch tip are equal. When the tips are molecular data (i.e., DNA, RNA and protein), the ultrametricity assumption is called the molecular clock.
Working example
First step
- First clustering
Let us assume that we have five elements  and the following matrix
and the following matrix  of pairwise distances between them :
of pairwise distances between them :
| a | b | c | d | e | |
|---|---|---|---|---|---|
| a | 0 | 17 | 21 | 31 | 23 |
| b | 17 | 0 | 30 | 34 | 21 |
| c | 21 | 30 | 0 | 28 | 39 |
| d | 31 | 34 | 28 | 0 | 43 |
| e | 23 | 21 | 39 | 43 | 0 |
In this example, 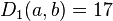 is the smallest value of
is the smallest value of  , so we join elements
, so we join elements 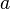 and
and  .
.
- First branch length estimation
Let 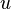 denote the node to which
denote the node to which 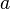 and
and  are now connected. Setting
are now connected. Setting  ensures that elements
ensures that elements 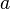 and
and  are equidistant from
are equidistant from 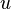 . This corresponds to the expectation of the ultrametricity hypothesis. The branches joining
. This corresponds to the expectation of the ultrametricity hypothesis. The branches joining 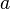 and
and  to
to 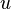 then have lengths
then have lengths  (see the final dendrogram)
(see the final dendrogram)
- First distance matrix update
We then proceed to update the initial distance matrix  into a new distance matrix
into a new distance matrix  (see below), reduced in size by one row and one column because of the clustering of
(see below), reduced in size by one row and one column because of the clustering of 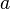 with
with  . Bold values in
. Bold values in  correspond to the new distances, calculated by averaging distances between the first cluster
correspond to the new distances, calculated by averaging distances between the first cluster  and each of the remaining elements:
and each of the remaining elements:

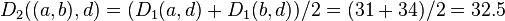
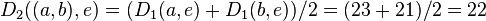
Italicized values in  are not affected by the matrix update as they correspond to distances between elements not involved in the first cluster.
are not affected by the matrix update as they correspond to distances between elements not involved in the first cluster.
Second step
- Second clustering
We now reiterate the three previous steps, starting from the new distance matrix 
| (a,b) | c | d | e | |
|---|---|---|---|---|
| (a,b) | 0 | 25.5 | 32.5 | 22 |
| c | 25.5 | 0 | 28 | 39 |
| d | 32.5 | 28 | 0 | 43 |
| e | 22 | 39 | 43 | 0 |
Here,  is the smallest value of
is the smallest value of  , so we join cluster
, so we join cluster  and element
and element  .
.
- Second branch length estimation
Let  denote the node to which
denote the node to which  and
and  are now connected. Because of the ultrametricity constraint, the branches joining
are now connected. Because of the ultrametricity constraint, the branches joining 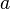 or
or  to
to  , and
, and  to
to  are equal and have the following length:
are equal and have the following length: 
We deduce the missing branch length:  (see the final dendrogram)
(see the final dendrogram)
- Second distance matrix update
We then proceed to update  into a new distance matrix
into a new distance matrix  (see below), reduced in size by one row and one column because of the clustering of
(see below), reduced in size by one row and one column because of the clustering of  with
with  . Bold values in
. Bold values in  correspond to the new distances, calculated by proportional averaging:
correspond to the new distances, calculated by proportional averaging:
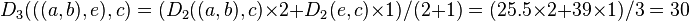
Thanks to this proportional average, the calculation of this new distance accounts for the larger size of the  cluster (two elements) with respect to
cluster (two elements) with respect to  (one element). Similarly:
(one element). Similarly:
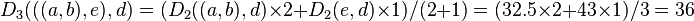
Proportional averaging therefore gives equal weight to the initial distances of matrix  . This is the reason why the method is unweighted, not with respect to the mathematical procedure but with respect to the initial distances.
. This is the reason why the method is unweighted, not with respect to the mathematical procedure but with respect to the initial distances.
Third step
- Third clustering
We again reiterate the three previous steps, starting from the updated distance matrix  .
.
| ((a,b),e) | c | d | |
|---|---|---|---|
| ((a,b),e) | 0 | 30 | 36 |
| c | 30 | 0 | 28 |
| d | 36 | 28 | 0 |
Here,  is the smallest value of
is the smallest value of  , so we join elements
, so we join elements  and
and 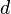 .
.
- Third branch length estimation
Let  denote the node to which
denote the node to which  and
and 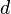 are now connected. The branches joining
are now connected. The branches joining  and
and 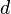 to
to  then have lengths
then have lengths  (see the final dendrogram)
(see the final dendrogram)
- Third distance matrix update
There is a single entry to update, keeping in mind that the two elements  and
and 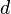 each have a contribution of
each have a contribution of  in the average computation:
in the average computation:

Final step
The final  matrix is:
matrix is:
| ((a,b),e) | (c,d) | |
|---|---|---|
| ((a,b),e) | 0 | 33 |
| (c,d) | 33 | 0 |
So we join clusters  and
and  .
.
Let  denote the (root) node to which
denote the (root) node to which  and
and  are now connected. The branches joining
are now connected. The branches joining  and
and  to
to  then have lengths:
then have lengths:
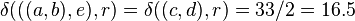
We deduce the two remaining branch lengths:


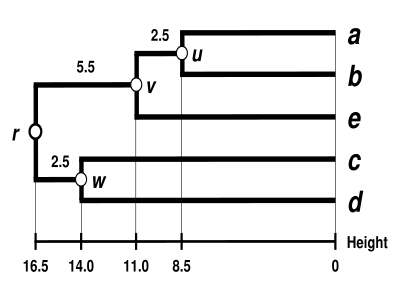
The UPGMA dendrogram
The dendrogram is now complete. It is ultrametric because all tips ( to
to  ) are equidistant from
) are equidistant from  :
:

The dendrogram is therefore rooted by  , its deepest node.
, its deepest node.
Uses
- In ecology, it is one of the most popular methods for the classification of sampling units (such as vegetation plots) on the basis of their pairwise similarities in relevant descriptor variables (such as species composition).[2]
- In bioinformatics, UPGMA is used for the creation of phenetic trees (phenograms). UPGMA was initially designed for use in protein electrophoresis studies, but is currently most often used to produce guide trees for more sophisticated algorithms. This algorithm is for example used in sequence alignment procedures, as it proposes one order in which the sequences will be aligned. Indeed, the guide tree aims at grouping the most similar sequences, regardless of their evolutionary rate or phylogenetic affinities, and that is exactly the goal of UPGMA.[3]
- In phylogenetics, UPGMA assumes a constant rate of evolution (molecular clock hypothesis), and is not a well-regarded method for inferring relationships unless this assumption has been tested and justified for the data set being used.
Time complexity
A trivial implementation of the algorithm to construct the UPGMA tree has  time complexity, and using a heap for each cluster to keep its distances from other cluster reduces its time to
time complexity, and using a heap for each cluster to keep its distances from other cluster reduces its time to 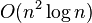 . Fionn Murtagh presented some other approaches for special cases, a
. Fionn Murtagh presented some other approaches for special cases, a  time algorithm by Day and Edelsbrunner[4] for k-dimensional data that is optimal
time algorithm by Day and Edelsbrunner[4] for k-dimensional data that is optimal 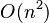 for constant k, and another
for constant k, and another  algorithm for restricted inputs, when "the anglomerative strategy satisfies the reducibility property."[5]
algorithm for restricted inputs, when "the anglomerative strategy satisfies the reducibility property."[5]






















 3362
3362

 被折叠的 条评论
为什么被折叠?
被折叠的 条评论
为什么被折叠?








