声明:内容非原创,是学习内容的总结,版权所属姜老师
线性回归引入
# 线性回归导入
from sklearn.linear_model import LinearRegression
from sklearn.neighbors import KNeighborsRegressor
import numpy as np
import pandas as pd
from pandas import Series,DataFrame
import matplotlib.pyplot as plt
%matplotlib inline
# 生成一组假数据 假设x和y都遵循正弦分布
x = np.linspace(0, 2*np.pi, 60)
y = np.sin(x)
plt.scatter(x,y)
<matplotlib.collections.PathCollection at 0x1facb5abdc0>
# 生成噪声数据 (-0.5, 0.5)
bias = np.random.random(30) - 0.5
bias
array([-0.39502105, 0.24340303, 0.08361402, -0.00268972, -0.03143423,
-0.41971947, 0.2567082 , -0.07361341, -0.14754526, -0.41319023,
0.21895744, -0.31080432, 0.21599872, -0.08704963, -0.1380524 ,
-0.23046208, 0.3592808 , -0.16425466, -0.00648164, 0.24217655,
0.43796813, 0.21696034, 0.4143489 , 0.22052109, 0.25845568,
-0.35609606, -0.00906317, -0.24740159, -0.21426299, -0.13506899])
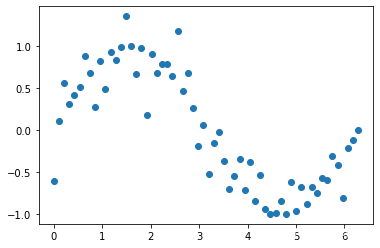
# 把噪声数据添加到y中
y[::2] += bias
plt.scatter(x,y)
<matplotlib.collections.PathCollection at 0x1facc2489a0>
x.shape
(60,)
# vsm 映射
X = x.reshape(-1,1)
X.shape
(60, 1)
X
array([[0. ],
[0.10649467],
[0.21298933],
[0.319484 ],
[0.42597866],
[0.53247333],
[0.638968 ],
[0.74546266],
[0.85195733],
[0.958452 ],
[1.06494666],
[1.17144133],
[1.27793599],
[1.38443066],
[1.49092533],
[1.59741999],
[1.70391466],
[1.81040933],
[1.91690399],
[2.02339866],
[2.12989332],
[2.23638799],
[2.34288266],
[2.44937732],
[2.55587199],
[2.66236666],
[2.76886132],
[2.87535599],
[2.98185065],
[3.08834532],
[3.19483999],
[3.30133465],
[3.40782932],
[3.51432399],
[3.62081865],
[3.72731332],
[3.83380798],
[3.94030265],
[4.04679732],
[4.15329198],
[4.25978665],
[4.36628132],
[4.47277598],
[4.57927065],
[4.68576531],
[4.79225998],
[4.89875465],
[5.00524931],
[5.11174398],
[5.21823864],
[5.32473331],
[5.43122798],
[5.53772264],
[5.64421731],
[5.75071198],
[5.85720664],
[5.96370131],
[6.07019597],
[6.17669064],
[6.28318531]])
# 1. knn实例化
knn =KNeighborsRegressor(n_neighbors=3)
# 2. 训练fit 实例化好的knn回归器
knn.fit(X, y)
KNeighborsRegressor(n_neighbors=3)
# 生成测试数据,别忘了和X同样的结构
# 注意,生成的数据只要不是60就行, 因为我们样本集数据也是同样范围的等差计算出来的
X_test = np.linspace(0, 2*np.pi, 45).reshape(-1,1)
X_test
array([[0. ],
[0.14279967],
[0.28559933],
[0.428399 ],
[0.57119866],
[0.71399833],
[0.856798 ],
[0.99959766],
[1.14239733],
[1.28519699],
[1.42799666],
[1.57079633],
[1.71359599],
[1.85639566],
[1.99919533],
[2.14199499],
[2.28479466],
[2.42759432],
[2.57039399],
[2.71319366],
[2.85599332],
[2.99879299],
[3.14159265],
[3.28439232],
[3.42719199],
[3.56999165],
[3.71279132],
[3.85559098],
[3.99839065],
[4.14119032],
[4.28398998],
[4.42678965],
[4.56958931],
[4.71238898],
[4.85518865],
[4.99798831],
[5.14078798],
[5.28358764],
[5.42638731],
[5.56918698],
[5.71198664],
[5.85478631],
[5.99758598],
[6.14038564],
[6.28318531]])
# 3.预测:使用X_test作为特征
y1_ = knn.predict(X_test)
y1_
array([ 0.01827837, 0.01827837, 0.42913475, 0.41198503, 0.60033228,
0.61071011, 0.59043822, 0.52817234, 0.74745077, 0.91117407,
1.05438104, 1.00544205, 0.87894106, 0.60410825, 0.58399583,
0.78779728, 0.74957241, 0.86489298, 0.75704464, 0.76885007,
0.24759279, 0.04078003, -0.21986071, -0.2357875 , -0.18423447,
-0.53796589, -0.53252107, -0.53902763, -0.64776802, -0.58808059,
-0.77527639, -0.9766071 , -0.94502224, -0.94463684, -0.85647279,
-0.75180678, -0.83833303, -0.76975748, -0.66825555, -0.6397318 ,
-0.43908767, -0.50822301, -0.47612273, -0.11130414, -0.11130414])
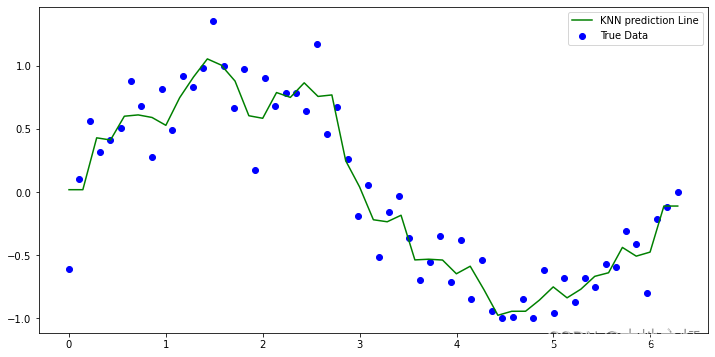
# 绘制 knn回归的预测结果
plt.figure(figsize=(12,6))
plt.scatter(x, y ,label='True Data', color ='blue')
plt.plot(X_test,y1_, label='KNN prediction Line',color = 'green')
plt.legend()
plt.show()
# 使用线性回归做预测
# 1.实例化线性模型 linearRegression
linear = LinearRegression()
# 2.fit训练模型 给定x和y
linear.fit(X,y)
LinearRegression()
# 3.模型预测 利用X_test进行预测
y2_ = linear.predict(X_test)
y2_
array([ 8.35573602e-01, 7.97617275e-01, 7.59660948e-01, 7.21704620e-01,
6.83748293e-01, 6.45791966e-01, 6.07835639e-01, 5.69879311e-01,
5.31922984e-01, 4.93966657e-01, 4.56010330e-01, 4.18054002e-01,
3.80097675e-01, 3.42141348e-01, 3.04185021e-01, 2.66228693e-01,
2.28272366e-01, 1.90316039e-01, 1.52359712e-01, 1.14403384e-01,
7.64470572e-02, 3.84907299e-02, 5.34402699e-04, -3.74219245e-02,
-7.53782518e-02, -1.13334579e-01, -1.51290906e-01, -1.89247234e-01,
-2.27203561e-01, -2.65159888e-01, -3.03116215e-01, -3.41072542e-01,
-3.79028870e-01, -4.16985197e-01, -4.54941524e-01, -4.92897851e-01,
-5.30854179e-01, -5.68810506e-01, -6.06766833e-01, -6.44723160e-01,
-6.82679488e-01, -7.20635815e-01, -7.58592142e-01, -7.96548469e-01,
-8.34504797e-01])
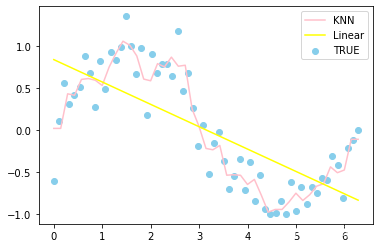
# 预测结果作一个绘制
plt.scatter(x,y,label='TRUE',color = 'skyblue')
plt.plot(X_test,y1_,label='KNN',color='pink')
plt.plot(X_test,y2_,label='Linear',color='yellow')
plt.legend()
plt.show()
knn线性回归(糖尿病数据集)
```python
import numpy as np
import pandas as pd
from pandas import Series,DataFrame
import matplotlib.pyplot as plt
import seaborn as sns
%matplotlib inline
from sklearn.linear_model import LinearRegression
from sklearn.neighbors import KNeighborsRegressor
from sklearn.datasets import load_diabetes
# 这个是diabetes糖尿病数据集
diabetes = load_diabetes()
diabetes
{'data': array([[ 0.03807591, 0.05068012, 0.06169621, ..., -0.00259226,
0.01990842, -0.01764613],
[-0.00188202, -0.04464164, -0.05147406, ..., -0.03949338,
-0.06832974, -0.09220405],
[ 0.08529891, 0.05068012, 0.04445121, ..., -0.00259226,
0.00286377, -0.02593034],
...,
[ 0.04170844, 0.05068012, -0.01590626, ..., -0.01107952,
-0.04687948, 0.01549073],
[-0.04547248, -0.04464164, 0.03906215, ..., 0.02655962,
0.04452837, -0.02593034],
[-0.04547248, -0.04464164, -0.0730303 , ..., -0.03949338,
-0.00421986, 0.00306441]]),
'target': array([151., 75., 141., 206., 135., 97., 138., 63., 110., 310., 101.,
69., 179., 185., 118., 171., 166., 144., 97., 168., 68., 49.,
68., 245., 184., 202., 137., 85., 131., 283., 129., 59., 341.,
87., 65., 102., 265., 276., 252., 90., 100., 55., 61., 92.,
259., 53., 190., 142., 75., 142., 155., 225., 59., 104., 182.,
128., 52., 37., 170., 170., 61., 144., 52., 128., 71., 163.,
150., 97., 160., 178., 48., 270., 202., 111., 85., 42., 170.,
200., 252., 113., 143., 51., 52., 210., 65., 141., 55., 134.,
42., 111., 98., 164., 48., 96., 90., 162., 150., 279., 92.,
83., 128., 102., 302., 198., 95., 53., 134., 144., 232., 81.,
104., 59., 246., 297., 258., 229., 275., 281., 179., 200., 200.,
173., 180., 84., 121., 161., 99., 109., 115., 268., 274., 158.,
107., 83., 103., 272., 85., 280., 336., 281., 118., 317., 235.,
60., 174., 259., 178., 128., 96., 126., 288., 88., 292., 71.,
197., 186., 25., 84., 96., 195., 53., 217., 172., 131., 214.,
59., 70., 220., 268., 152., 47., 74., 295., 101., 151., 127.,
237., 225., 81., 151., 107., 64., 138., 185., 265., 101., 137.,
143., 141., 79., 292., 178., 91., 116., 86., 122., 72., 129.,
142., 90., 158., 39., 196., 222., 277., 99., 196., 202., 155.,
77., 191., 70., 73., 49., 65., 263., 248., 296., 214., 185.,
78., 93., 252., 150., 77., 208., 77., 108., 160., 53., 220.,
154., 259., 90., 246., 124., 67., 72., 257., 262., 275., 177.,
71., 47., 187., 125., 78., 51., 258., 215., 303., 243., 91.,
150., 310., 153., 346., 63., 89., 50., 39., 103., 308., 116.,
145., 74., 45., 115., 264., 87., 202., 127., 182., 241., 66.,
94., 283., 64., 102., 200., 265., 94., 230., 181., 156., 233.,
60., 219., 80., 68., 332., 248., 84., 200., 55., 85., 89.,
31., 129., 83., 275., 65., 198., 236., 253., 124., 44., 172.,
114., 142., 109., 180., 144., 163., 147., 97., 220., 190., 109.,
191., 122., 230., 242., 248., 249., 192., 131., 237., 78., 135.,
244., 199., 270., 164., 72., 96., 306., 91., 214., 95., 216.,
263., 178., 113., 200., 139., 139., 88., 148., 88., 243., 71.,
77., 109., 272., 60., 54., 221., 90., 311., 281., 182., 321.,
58., 262., 206., 233., 242., 123., 167., 63., 197., 71., 168.,
140., 217., 121., 235., 245., 40., 52., 104., 132., 88., 69.,
219., 72., 201., 110., 51., 277., 63., 118., 69., 273., 258.,
43., 198., 242., 232., 175., 93., 168., 275., 293., 281., 72.,
140., 189., 181., 209., 136., 261., 113., 131., 174., 257., 55.,
84., 42., 146., 212., 233., 91., 111., 152., 120., 67., 310.,
94., 183., 66., 173., 72., 49., 64., 48., 178., 104., 132.,
220., 57.]),
'frame': None,
'DESCR': '.. _diabetes_dataset:\n\nDiabetes dataset\n----------------\n\nTen baseline variables, age, sex, body mass index, average blood\npressure, and six blood serum measurements were obtained for each of n =\n442 diabetes patients, as well as the response of interest, a\nquantitative measure of disease progression one year after baseline.\n\n**Data Set Characteristics:**\n\n :Number of Instances: 442\n\n :Number of Attributes: First 10 columns are numeric predictive values\n\n :Target: Column 11 is a quantitative measure of disease progression one year after baseline\n\n :Attribute Information:\n - age age in years\n - sex\n - bmi body mass index\n - bp average blood pressure\n - s1 tc, total serum cholesterol\n - s2 ldl, low-density lipoproteins\n - s3 hdl, high-density lipoproteins\n - s4 tch, total cholesterol / HDL\n - s5 ltg, possibly log of serum triglycerides level\n - s6 glu, blood sugar level\n\nNote: Each of these 10 feature variables have been mean centered and scaled by the standard deviation times `n_samples` (i.e. the sum of squares of each column totals 1).\n\nSource URL:\nhttps://www4.stat.ncsu.edu/~boos/var.select/diabetes.html\n\nFor more information see:\nBradley Efron, Trevor Hastie, Iain Johnstone and Robert Tibshirani (2004) "Least Angle Regression," Annals of Statistics (with discussion), 407-499.\n(https://web.stanford.edu/~hastie/Papers/LARS/LeastAngle_2002.pdf)',
'feature_names': ['age',
'sex',
'bmi',
'bp',
's1',
's2',
's3',
's4',
's5',
's6'],
'data_filename': 'D:\\software\\anaconda\\lib\\site-packages\\sklearn\\datasets\\data\\diabetes_data.csv.gz',
'target_filename': 'D:\\software\\anaconda\\lib\\site-packages\\sklearn\\datasets\\data\\diabetes_target.csv.gz'}
print(diabetes.DESCR)
.. _diabetes_dataset:
Diabetes dataset
----------------
Ten baseline variables, age, sex, body mass index, average blood
pressure, and six blood serum measurements were obtained for each of n =
442 diabetes patients, as well as the response of interest, a
quantitative measure of disease progression one year after baseline.
**Data Set Characteristics:**
:Number of Instances: 442
:Number of Attributes: First 10 columns are numeric predictive values
:Target: Column 11 is a quantitative measure of disease progression one year after baseline
:Attribute Information:
- age age in years
- sex
- bmi body mass index
- bp average blood pressure
- s1 tc, total serum cholesterol
- s2 ldl, low-density lipoproteins
- s3 hdl, high-density lipoproteins
- s4 tch, total cholesterol / HDL
- s5 ltg, possibly log of serum triglycerides level
- s6 glu, blood sugar level
Note: Each of these 10 feature variables have been mean centered and scaled by the standard deviation times `n_samples` (i.e. the sum of squares of each column totals 1).
Source URL:
https://www4.stat.ncsu.edu/~boos/var.select/diabetes.html
For more information see:
Bradley Efron, Trevor Hastie, Iain Johnstone and Robert Tibshirani (2004) "Least Angle Regression," Annals of Statistics (with discussion), 407-499.
(https://web.stanford.edu/~hastie/Papers/LARS/LeastAngle_2002.pdf)
diabetes.feature_names
['age', 'sex', 'bmi', 'bp', 's1', 's2', 's3', 's4', 's5', 's6']
# 导出数据成为DataFrame
dataSets = DataFrame(data=diabetes.data,columns=diabetes.feature_names)
dataSets.head()
| age | sex | bmi | bp | s1 | s2 | s3 | s4 | s5 | s6 | |
|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 0.038076 | 0.050680 | 0.061696 | 0.021872 | -0.044223 | -0.034821 | -0.043401 | -0.002592 | 0.019908 | -0.017646 |
| 1 | -0.001882 | -0.044642 | -0.051474 | -0.026328 | -0.008449 | -0.019163 | 0.074412 | -0.039493 | -0.068330 | -0.092204 |
| 2 | 0.085299 | 0.050680 | 0.044451 | -0.005671 | -0.045599 | -0.034194 | -0.032356 | -0.002592 | 0.002864 | -0.025930 |
| 3 | -0.089063 | -0.044642 | -0.011595 | -0.036656 | 0.012191 | 0.024991 | -0.036038 | 0.034309 | 0.022692 | -0.009362 |
| 4 | 0.005383 | -0.044642 | -0.036385 | 0.021872 | 0.003935 | 0.015596 | 0.008142 | -0.002592 | -0.031991 | -0.046641 |
dataSets.describe().T
| count | mean | std | min | 25% | 50% | 75% | max | |
|---|---|---|---|---|---|---|---|---|
| age | 442.0 | -3.634285e-16 | 0.047619 | -0.107226 | -0.037299 | 0.005383 | 0.038076 | 0.110727 |
| sex | 442.0 | 1.308343e-16 | 0.047619 | -0.044642 | -0.044642 | -0.044642 | 0.050680 | 0.050680 |
| bmi | 442.0 | -8.045349e-16 | 0.047619 | -0.090275 | -0.034229 | -0.007284 | 0.031248 | 0.170555 |
| bp | 442.0 | 1.281655e-16 | 0.047619 | -0.112400 | -0.036656 | -0.005671 | 0.035644 | 0.132044 |
| s1 | 442.0 | -8.835316e-17 | 0.047619 | -0.126781 | -0.034248 | -0.004321 | 0.028358 | 0.153914 |
| s2 | 442.0 | 1.327024e-16 | 0.047619 | -0.115613 | -0.030358 | -0.003819 | 0.029844 | 0.198788 |
| s3 | 442.0 | -4.574646e-16 | 0.047619 | -0.102307 | -0.035117 | -0.006584 | 0.029312 | 0.181179 |
| s4 | 442.0 | 3.777301e-16 | 0.047619 | -0.076395 | -0.039493 | -0.002592 | 0.034309 | 0.185234 |
| s5 | 442.0 | -3.830854e-16 | 0.047619 | -0.126097 | -0.033249 | -0.001948 | 0.032433 | 0.133599 |
| s6 | 442.0 | -3.412882e-16 | 0.047619 | -0.137767 | -0.033179 | -0.001078 | 0.027917 | 0.135612 |
# 样本集拆分
from sklearn.model_selection import train_test_split
# 标签拿进来
target = diabetes.target
# 拆分训练集测试集
X_train,X_test,y_train,y_test = train_test_split(dataSets, target,test_size=0.2, random_state=2)
# 1.实例化模型 knn
knn = KNeighborsRegressor()
# 2.训练模型fit
knn.fit(X_train,y_train)
KNeighborsRegressor()
# MAE MSE
from sklearn.metrics import mean_squared_error, mean_absolute_error
# 泛化误差 -mse
mean_squared_error(y_test, knn.predict(X_test))
3941.039550561798
# 平均绝对误差 mae
mean_absolute_error(y_test, knn.predict(X_test))
50.06966292134831
target
array([151., 75., 141., 206., 135., 97., 138., 63., 110., 310., 101.,
69., 179., 185., 118., 171., 166., 144., 97., 168., 68., 49.,
68., 245., 184., 202., 137., 85., 131., 283., 129., 59., 341.,
87., 65., 102., 265., 276., 252., 90., 100., 55., 61., 92.,
259., 53., 190., 142., 75., 142., 155., 225., 59., 104., 182.,
128., 52., 37., 170., 170., 61., 144., 52., 128., 71., 163.,
150., 97., 160., 178., 48., 270., 202., 111., 85., 42., 170.,
200., 252., 113., 143., 51., 52., 210., 65., 141., 55., 134.,
42., 111., 98., 164., 48., 96., 90., 162., 150., 279., 92.,
83., 128., 102., 302., 198., 95., 53., 134., 144., 232., 81.,
104., 59., 246., 297., 258., 229., 275., 281., 179., 200., 200.,
173., 180., 84., 121., 161., 99., 109., 115., 268., 274., 158.,
107., 83., 103., 272., 85., 280., 336., 281., 118., 317., 235.,
60., 174., 259., 178., 128., 96., 126., 288., 88., 292., 71.,
197., 186., 25., 84., 96., 195., 53., 217., 172., 131., 214.,
59., 70., 220., 268., 152., 47., 74., 295., 101., 151., 127.,
237., 225., 81., 151., 107., 64., 138., 185., 265., 101., 137.,
143., 141., 79., 292., 178., 91., 116., 86., 122., 72., 129.,
142., 90., 158., 39., 196., 222., 277., 99., 196., 202., 155.,
77., 191., 70., 73., 49., 65., 263., 248., 296., 214., 185.,
78., 93., 252., 150., 77., 208., 77., 108., 160., 53., 220.,
154., 259., 90., 246., 124., 67., 72., 257., 262., 275., 177.,
71., 47., 187., 125., 78., 51., 258., 215., 303., 243., 91.,
150., 310., 153., 346., 63., 89., 50., 39., 103., 308., 116.,
145., 74., 45., 115., 264., 87., 202., 127., 182., 241., 66.,
94., 283., 64., 102., 200., 265., 94., 230., 181., 156., 233.,
60., 219., 80., 68., 332., 248., 84., 200., 55., 85., 89.,
31., 129., 83., 275., 65., 198., 236., 253., 124., 44., 172.,
114., 142., 109., 180., 144., 163., 147., 97., 220., 190., 109.,
191., 122., 230., 242., 248., 249., 192., 131., 237., 78., 135.,
244., 199., 270., 164., 72., 96., 306., 91., 214., 95., 216.,
263., 178., 113., 200., 139., 139., 88., 148., 88., 243., 71.,
77., 109., 272., 60., 54., 221., 90., 311., 281., 182., 321.,
58., 262., 206., 233., 242., 123., 167., 63., 197., 71., 168.,
140., 217., 121., 235., 245., 40., 52., 104., 132., 88., 69.,
219., 72., 201., 110., 51., 277., 63., 118., 69., 273., 258.,
43., 198., 242., 232., 175., 93., 168., 275., 293., 281., 72.,
140., 189., 181., 209., 136., 261., 113., 131., 174., 257., 55.,
84., 42., 146., 212., 233., 91., 111., 152., 120., 67., 310.,
94., 183., 66., 173., 72., 49., 64., 48., 178., 104., 132.,
220., 57.])
# 消除数据的量级(量纲)
# 1. 标准化 Z-SCORE StanderdScaller - 均值为0 标准差为1
# 2. 区缩放 MinMaxScaller 缩放到[0,1]
from sklearn.preprocessing import StandardScaler, MinMaxScaler
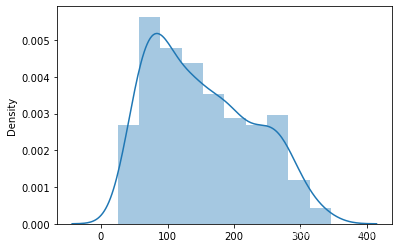
# 首先 查看target的数据分布
sns.distplot(target)
<AxesSubplot:ylabel='Density'>
# 1. 实例化
ss = StandardScaler()
# 2. 转化
ss_target = ss.fit_transform(target.reshape(-1,1))
ss_target
array([[-1.47194752e-02],
[-1.00165882e+00],
[-1.44579915e-01],
[ 6.99512942e-01],
[-2.22496178e-01],
[-7.15965848e-01],
[-1.83538046e-01],
[-1.15749134e+00],
[-5.47147277e-01],
[ 2.05006151e+00],
......
[-2.61454310e-01],
[ 8.81317557e-01],
[-1.23540761e+00]])
sns.distplot(ss_target)
<AxesSubplot:ylabel='Density'>
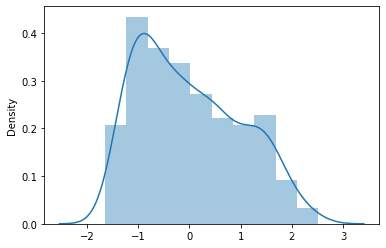
# 使用标准化之后的target 来建模(ss_target)
# 【注意】 random_state 要和之前的保持一致, 为的是保证两个模型使用的训练集和测试集一致
X_train, X_test, y_train, y_test = train_test_split(dataSets, ss_target, test_size=0.2, random_state=2)
# 重新训练k=5这个对象
knn.fit(X_train,y_train)
KNeighborsRegressor()
# 训练之后的新的评判
mean_squared_error(y_test,knn.predict(X_test))
0.6646064163260871
mean_absolute_error(y_test,knn.predict(X_test))
0.6502068430871709
找到一个更好的模型(knn调参,评价(MAE,MSE))
# 先试一下使用线性回归
#1.实例化
linear = LinearRegression()
#2.训
linear.fit(X_train,y_train)
#3.评判
mean_squared_error(y_test,linear.predict(X_test))
0.5218363683134858
问题:
- knn是可以调参的
- 算法评分是某一次的随机值
- 回归问题模型的好坏,要看泛化误差 和 经验误差
# 定义一个函数来评判模型
def cal_score(model, X,y, count, test_size):
train_mse_list= []
test_mse_list = []
for i in range(count):
X_train,X_test,y_train,y_test = train_test_split(X,y,test_size=test_size)
model.fit(X,y)
train_mse_list.append(mean_squared_error(y_train, model.predict(X_train)))
test_mse_list.append(mean_squared_error(y_test, model.predict(X_test)))
return np.array(train_mse_list),np.array(test_mse_list)
knn = KNeighborsRegressor(n_neighbors=5)
train_score_array,test_score_array = cal_score(model=knn,X=dataSets.values,y=target,count=10,test_size=0.3)
train_score_array.mean() ,train_score_array.std()
(2336.3235339805824, 87.16520782728838)
test_score_array.mean() ,test_score_array.std()
(2357.03569924812, 202.51164826039167)
linear = LinearRegression()
train_score_array,test_score_array = cal_score(model=linear,X=dataSets.values,y=target,count=10,test_size=0.3)
train_score_array.mean() ,train_score_array.std()
(2807.125616206974, 100.88450030074503)
test_score_array.mean() ,test_score_array.std()
(2981.8145928385716, 234.3857939318062)
# 评判一下 在不同的k值上的knn模型的表现
k_list = np.arange(1,21,2)
cols = []
for k in k_list:
knn = KNeighborsRegressor(n_neighbors=k)
train_scores,test_scores = cal_score(model=knn,X=dataSets.values,y=target,count=10,test_size=0.2)
row = [k,
train_scores.mean().tolist(),
test_scores.mean().tolist(),
train_scores.std().tolist(),
test_scores.std().tolist()
]
cols.append(row)
result= DataFrame(data=np.array(cols),columns=['k','Train_mean','Test_mean','Train_std','Test_std'])
result
| k | Train_mean | Test_mean | Train_std | Test_std | |
|---|---|---|---|---|---|
| 0 | 1.0 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |
| 1 | 3.0 | 1937.297136 | 2021.645443 | 72.113122 | 286.021709 |
| 2 | 5.0 | 2333.390946 | 2378.906921 | 111.330407 | 441.568918 |
| 3 | 7.0 | 2635.195207 | 2542.229420 | 52.116173 | 206.707967 |
| 4 | 9.0 | 2731.609684 | 2615.471952 | 72.131433 | 286.094334 |
| 5 | 11.0 | 2724.262086 | 2998.797892 | 59.500027 | 235.994490 |
| 6 | 13.0 | 2829.240362 | 2869.213197 | 40.296168 | 159.826374 |
| 7 | 15.0 | 2794.546657 | 3125.196095 | 68.837757 | 273.030655 |
| 8 | 17.0 | 2898.627282 | 2875.761736 | 69.569247 | 275.931959 |
| 9 | 19.0 | 2868.687365 | 2924.798718 | 66.040720 | 261.936788 |
result.set_index('k').plot()
plt.xticks(result['k'].values)
plt.show()
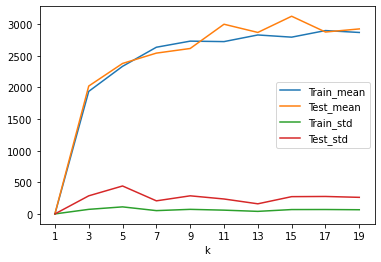
# 1.经验误差和泛化误差越接近并且越小
# 2. 经验误差和泛化误差的标准方差更小,越小越稳定
# 我们还能做什么?
# 还可以做特征处理。
# 理解数据
dataSets.describe().T
| count | mean | std | min | 25% | 50% | 75% | max | |
|---|---|---|---|---|---|---|---|---|
| age | 442.0 | -3.634285e-16 | 0.047619 | -0.107226 | -0.037299 | 0.005383 | 0.038076 | 0.110727 |
| sex | 442.0 | 1.308343e-16 | 0.047619 | -0.044642 | -0.044642 | -0.044642 | 0.050680 | 0.050680 |
| bmi | 442.0 | -8.045349e-16 | 0.047619 | -0.090275 | -0.034229 | -0.007284 | 0.031248 | 0.170555 |
| bp | 442.0 | 1.281655e-16 | 0.047619 | -0.112400 | -0.036656 | -0.005671 | 0.035644 | 0.132044 |
| s1 | 442.0 | -8.835316e-17 | 0.047619 | -0.126781 | -0.034248 | -0.004321 | 0.028358 | 0.153914 |
| s2 | 442.0 | 1.327024e-16 | 0.047619 | -0.115613 | -0.030358 | -0.003819 | 0.029844 | 0.198788 |
| s3 | 442.0 | -4.574646e-16 | 0.047619 | -0.102307 | -0.035117 | -0.006584 | 0.029312 | 0.181179 |
| s4 | 442.0 | 3.777301e-16 | 0.047619 | -0.076395 | -0.039493 | -0.002592 | 0.034309 | 0.185234 |
| s5 | 442.0 | -3.830854e-16 | 0.047619 | -0.126097 | -0.033249 | -0.001948 | 0.032433 | 0.133599 |
| s6 | 442.0 | -3.412882e-16 | 0.047619 | -0.137767 | -0.033179 | -0.001078 | 0.027917 | 0.135612 |
# 通过画图来查看数据分布
for col_name in dataSets.columns:
col_data = dataSets[col_name]
sns.distplot(col_data)
plt.title(col_name)
plt.show()
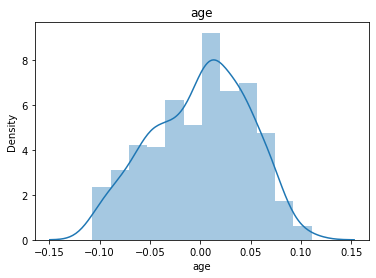
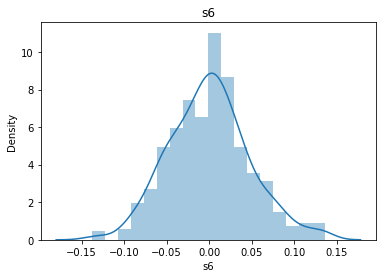
# 各种转化的目的 为了让特征值尽可能的正态分布
# 好处: 1.减少算法的迭代次数 2.数据降噪
岭回归
X W y
1 2 3 w1 1
2 3 4 w2 3
2 4 6 w3 2
a1 + b2 + c*3 =1
任何样本数据都必然拥有以下特点:
- 1. 存在噪声
- 2. 多重共线性
- 可能无法避免
如何解决这些问题呢: 角度一:算法的角度(正则项) 角度二:从数据集的角度(主要解决思路 也最有效果)
X.T *X
X W y
1+λ 2 3 w1 1
2 3+λ 4 w2 3
2 4 6+λ w3 2
y = f(x) + bias + 正则项
y = f(x) + 1 - 1
缩减算法:减少 无用特征影响系数w —>0
过拟合:模型对训练集的局部特征过度关注导致的
100收入 4身高 1地域 0.0005电话 0.01学历
岭回归的基本使用
import numpy as np
X = np.array([[1,2,3,4],[1,3,8,5]])
X
array([[1, 2, 3, 4],
[1, 3, 8, 5]])
y = np.array([3,5])
y
array([3, 5])
from sklearn.linear_model import LinearRegression,Ridge
lr = LinearRegression()
lr.fit(X, y)
LinearRegression()
# 岭回归的使用
ridge = Ridge()
ridge.fit(X,y)
Ridge()
获取线性回归的系数
lr.coef_
array([5.55111512e-17, 7.40740741e-02, 3.70370370e-01, 7.40740741e-02])
ridge.coef_
array([0. , 0.06896552, 0.34482759, 0.06896552])
获取线性回归的截距
lr.intercept_
1.4444444444444446
ridge.intercept_
1.6206896551724141
糖尿病的回归分析
from sklearn.datasets import load_diabetes
import pandas as pd
from pandas import Series,DataFrame
import matplotlib.pyplot as plt
%matplotlib inline
from sklearn.model_selection import train_test_split
# 拿出数据
diabetes = load_diabetes()
train = diabetes.data # X
target = diabetes.target # y
feature_names = diabetes.feature_names
feature_names
['age', 'sex', 'bmi', 'bp', 's1', 's2', 's3', 's4', 's5', 's6']
# 1.实例化岭回归
ridge = Ridge(alpha=1.0)
# 2.拆分测试机和训练集
X_train, X_test, y_train, y_test = train_test_split(train,target,test_size=0.2)
# 3.模型训练
ridge.fit(X_train,y_train)
Ridge()
利用系数来表达各个特征对目标的影响,进而可以根据这个影响的大小来进行特征选择
ridge.coef_
array([ 36.01468243, -90.75889609, 287.47200232, 180.86788548,
18.90897004, -23.19316941, -137.06157738, 123.45193373,
260.76150489, 110.61743971])
feature_names
['age', 'sex', 'bmi', 'bp', 's1', 's2', 's3', 's4', 's5', 's6']
import seaborn as sns
sns.set()
# 绘图来查看一下影响因子
importances = Series(data=ridge.coef_,index=feature_names)
importances.plot(kind='bar')
plt.xticks(rotation=0)
plt.show()
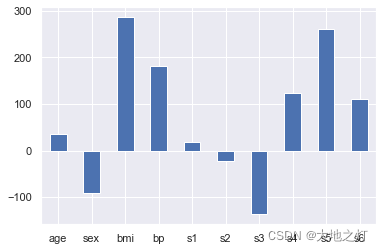
# 在特征重要性选择中,即使负数也会影响到整体,因此我们使用绝对值进行处理
importances = Series(data=np.abs(ridge.coef_),index=feature_names).sort_values(ascending=False)
importances.plot(kind='bar')
plt.xticks(rotation=0)
plt.show()
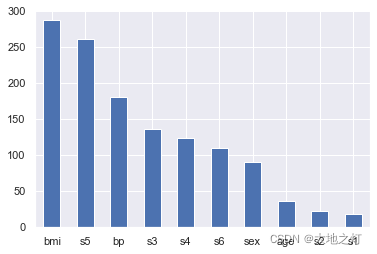
根据特征的重要性,我们发现前7个特征更加重要,因此我们可以选择前7个特征重新建模
importances
bmi 287.472002
s5 260.761505
bp 180.867885
s3 137.061577
s4 123.451934
s6 110.617440
sex 90.758896
age 36.014682
s2 23.193169
s1 18.908970
dtype: float64
importances_columns = ['bmi','s5','bp','s3','s4','s6','sex']
importances_columns
['bmi', 's5', 'bp', 's3', 's4', 's6', 'sex']
dataSets = DataFrame(data=train,columns=feature_names)
dataSets.head()
| age | sex | bmi | bp | s1 | s2 | s3 | s4 | s5 | s6 | |
|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 0.038076 | 0.050680 | 0.061696 | 0.021872 | -0.044223 | -0.034821 | -0.043401 | -0.002592 | 0.019908 | -0.017646 |
| 1 | -0.001882 | -0.044642 | -0.051474 | -0.026328 | -0.008449 | -0.019163 | 0.074412 | -0.039493 | -0.068330 | -0.092204 |
| 2 | 0.085299 | 0.050680 | 0.044451 | -0.005671 | -0.045599 | -0.034194 | -0.032356 | -0.002592 | 0.002864 | -0.025930 |
| 3 | -0.089063 | -0.044642 | -0.011595 | -0.036656 | 0.012191 | 0.024991 | -0.036038 | 0.034309 | 0.022692 | -0.009362 |
| 4 | 0.005383 | -0.044642 | -0.036385 | 0.021872 | 0.003935 | 0.015596 | 0.008142 | -0.002592 | -0.031991 | -0.046641 |
X = dataSets[importances_columns]
X.head()
| bmi | s5 | bp | s3 | s4 | s6 | sex | |
|---|---|---|---|---|---|---|---|
| 0 | 0.061696 | 0.019908 | 0.021872 | -0.043401 | -0.002592 | -0.017646 | 0.050680 |
| 1 | -0.051474 | -0.068330 | -0.026328 | 0.074412 | -0.039493 | -0.092204 | -0.044642 |
| 2 | 0.044451 | 0.002864 | -0.005671 | -0.032356 | -0.002592 | -0.025930 | 0.050680 |
| 3 | -0.011595 | 0.022692 | -0.036656 | -0.036038 | 0.034309 | -0.009362 | -0.044642 |
| 4 | -0.036385 | -0.031991 | 0.021872 | 0.008142 | -0.002592 | -0.046641 | -0.044642 |
y = target
# 已经做了特征选择的数据 重新进行划分
X_train, X_test, y_train,y_test = train_test_split(X,y,test_size=0.2,random_state=1)
# 对已经使用了特征选择的数据 进行建模
ridge = Ridge(alpha=1.0)
ridge.fit(X_train,y_train)
Ridge()
from sklearn.metrics import mean_squared_error
mean_squared_error(y_test,ridge.predict(X_test))
3276.750176885635
linear = LinearRegression()
linear.fit(X_train,y_train)
LinearRegression()
mean_squared_error(y_test,linear.predict(X_test))
3080.257052972186
# 比较 没有做特征选择的样本集
X_train1,X_test1,y_train1,y_test1 = train_test_split(train,target,test_size=0.2,random_state=1)
linear = LinearRegression()
linear.fit(X_train1,y_train1)
mean_squared_error(y_test1, linear.predict(X_test1))
2992.5576814529445
# Ridge LinearRegression 的系数都可以用来做特征选择
建模的核心:
数据质量(特征工程)
其次 才是算法和调参
# 特征工程 (目的:为了让模型可以得到更好的一个数据集)
# 特征选择 (基于算法的选择 常用的算法 基于线性的 决策树的..)
# 特征提取 (基于经验提取)
# 无量纲处理 (基于标准正态分布)
# 分箱操作
# 空值填充
# 异常值过滤
lasso回归
from sklearn.linear_model import Lasso,Ridge,LinearRegression
import numpy as np
X = np.array([[1,2,3,4,5],[2,3,2,1,7]])
X
array([[1, 2, 3, 4, 5],
[2, 3, 2, 1, 7]])
y = np.array([1,2])
y
array([1, 2])
# 使用拉索回归建模
# 1.实例化
lasso = Lasso(alpha=0.001)
# 2.训练模型
lasso.fit(X,y)
Lasso(alpha=0.001)
lasso.coef_
array([ 0. , 0. , -0. , -0.33288889, 0. ])
ridge = Ridge()
ridge.fit(X,y)
ridge.coef_
array([ 0.05555556, 0.05555556, -0.05555556, -0.16666667, 0.11111111])
linear = LinearRegression()
linear.fit(X,y)
linear.coef_
array([ 0.0625, 0.0625, -0.0625, -0.1875, 0.125 ])
三种不同的回归系数比较
a = np.array([1,2,3,4,5])
a
array([1, 2, 3, 4, 5])
a[[0,2]]= 0
a
array([0, 2, 0, 4, 5])
W[np.random.permutation(200)[10:]]
array([9.96516152e-01, 5.32391471e-01, 4.93905634e-02, 9.85577787e-01,
8.99366918e-01, 7.54742781e-01, 3.64525327e-01, 9.63422337e-01,
5.36894360e-01, 3.38068592e-01, 1.01662067e-01, 7.71842842e-01,
3.54289304e-01, 4.07413512e-01, 8.47113116e-01, 5.77998640e-01,
3.53312178e-01, 3.77569649e-02, 5.13445478e-01, 6.01971296e-01,
2.17216303e-01, 6.06875883e-01, 1.16501776e-01, 9.29483618e-01,
1.94729718e-01, 3.79243355e-01, 2.32664998e-01, 8.11550904e-01,
3.45585212e-01, 8.97118361e-01, 5.18677931e-01, 4.76157464e-02,
5.46642599e-01, 6.28107676e-01, 6.99770540e-01, 5.53885482e-01,
1.06766013e-01, 7.41079780e-01, 9.47096359e-01, 7.76124225e-02,
4.32180516e-01, 7.04356613e-01, 8.82416582e-01, 8.81437686e-02,
4.85216991e-02, 4.52000028e-01, 3.82505394e-01, 9.36136345e-01,
5.70745661e-01, 2.92274222e-01, 4.26258679e-01, 6.16950539e-01,
5.18476200e-01, 8.96571765e-01, 3.28256203e-01, 7.68933452e-01,
1.20772148e-01, 1.69006603e-02, 4.60183339e-01, 3.28257762e-01,
3.98607160e-01, 8.70438678e-01, 1.51239976e-01, 7.40343824e-01,
4.63024024e-01, 5.76156560e-01, 1.92047942e-03, 9.62859485e-01,
7.33956356e-01, 2.39740967e-01, 1.83232929e-01, 1.00671402e-01,
1.30679135e-01, 3.73608097e-01, 6.00838009e-01, 4.40710351e-01,
5.70835282e-01, 2.32309233e-01, 8.87733637e-01, 6.36192443e-01,
7.90478439e-01, 2.63124712e-01, 2.88636831e-01, 2.55830360e-02,
4.13459126e-01, 9.72222742e-01, 6.12679019e-01, 6.05975771e-01,
9.03416989e-01, 3.47247811e-01, 9.51756590e-01, 5.69286087e-01,
9.65890196e-01, 5.82139890e-01, 5.51093440e-01, 5.56825893e-01,
6.24669833e-01, 5.51314324e-01, 6.48828274e-01, 3.02442666e-01,
6.84657374e-01, 3.71998163e-01, 7.16317195e-01, 9.65935762e-01,
7.65903638e-01, 3.70943741e-01, 1.16563475e-01, 1.72965641e-01,
9.18789086e-01, 8.81839640e-01, 7.13009929e-01, 6.42216668e-01,
4.15987478e-01, 7.91627848e-02, 8.95408244e-01, 4.77017976e-01,
8.99848891e-01, 9.69446936e-01, 9.39478644e-01, 1.77600703e-01,
1.59593839e-01, 6.76554096e-01, 2.64041902e-01, 9.89595420e-01,
2.90237110e-01, 4.25976169e-01, 8.44444767e-01, 2.10848895e-02,
7.76392532e-01, 2.79007862e-01, 5.35291152e-02, 9.54334908e-01,
8.78255077e-01, 2.58898672e-01, 3.17425087e-01, 3.63382801e-01,
2.19014741e-01, 1.82570099e-01, 6.56801360e-01, 6.86939280e-01,
1.08914417e-01, 1.94762611e-04, 2.22156323e-01, 3.97132753e-01,
4.11637144e-01, 3.98905184e-01, 4.26635545e-01, 1.26789802e-01,
4.30105437e-02, 9.35596787e-01, 3.33434333e-01, 4.57113238e-01,
7.28383428e-01, 7.32163308e-01, 3.15656355e-02, 2.87251282e-01,
9.53105336e-01, 6.89125739e-01, 2.34995553e-01, 9.40084446e-01,
9.13471128e-01, 3.74629473e-01, 5.50133348e-01, 4.42872398e-01,
6.28822215e-01, 6.61706633e-01, 6.25204117e-01, 5.80891452e-01,
6.96894553e-01, 3.42318889e-01, 3.35023780e-01, 7.54764315e-01,
5.51524765e-01, 7.14882962e-01, 7.97392627e-01, 8.60508187e-01,
4.49895726e-01, 9.66104169e-01, 3.80176775e-01, 6.78332181e-01,
6.38685181e-01, 8.87437347e-01, 5.55300557e-01, 6.84089183e-01,
4.23983590e-01, 9.48933675e-01, 5.42585937e-01, 6.54043901e-01,
2.96119799e-01, 2.78872800e-01])
# 数据构造
samples = 50
features = 200
# 假设真实的数据中,对结果有真实作用的特征就10个
X = np.random.random(size=(samples,features))
W = np.random.random(features)
# W 这两百个数据中(系数),只有随机的10个是非0 其他都是0
W[np.random.permutation(200)[10:]] = 0
y = np.dot(X,W)
W
array([0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0.4271518 , 0. ,
0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. , 0.92177717,
0. , 0. , 0. , 0. , 0.31777111,
0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. , 0.55356324,
0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0.56237781, 0. ,
0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0.51434962, 0. , 0. ,
0. , 0. , 0.56461673, 0. , 0. ,
0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. , 0.18319368,
0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0.65038963, 0. , 0. ,
0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0.92661222, 0. , 0. ,
0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. , 0. ])
import matplotlib.pyplot as plt
%matplotlib inline
plt.plot(W)
[<matplotlib.lines.Line2D at 0x1c8ea5fcf40>]
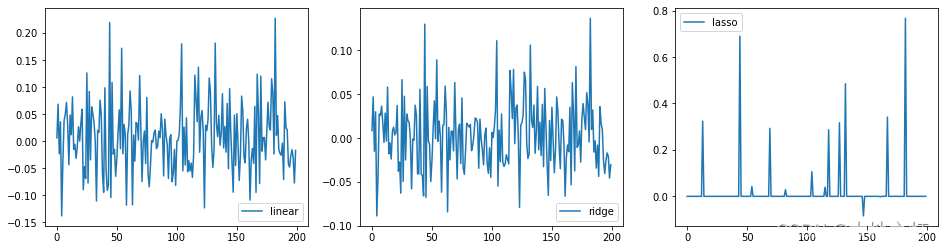
# 针对实战情况,三种不同的回归对比
# 1.实例化
linear = LinearRegression()
ridge = Ridge(alpha=10)
lasso = Lasso(alpha=0.01)
# 2.训练
linear.fit(X,y)
ridge.fit(X,y)
lasso.fit(X,y)
# 画图
plt.figure(figsize=(16,4))
ax1 = plt.subplot(1, 3, 1)
plt.plot(linear.coef_,label='linear')
plt.legend()
ax2 = plt.subplot(1,3,2)
plt.plot(ridge.coef_,label ='ridge')
plt.legend()
ax3 = plt.subplot(1,3,3)
plt.plot(lasso.coef_,label='lasso')
plt.legend()
<matplotlib.legend.Legend at 0x1c8ea6b7b50>





















 2956
2956











 被折叠的 条评论
为什么被折叠?
被折叠的 条评论
为什么被折叠?








