Before
Link to this article .pdf file
HW2: Numpy for Multi-Layer Neural Network
Refer to the answer:http://t.csdn.cn/9sdBN
Provide finished code files:
Get Files:
link:https://pan.baidu.com/s/1Fw_7thL5PxR79zI6XbpnYQ
password:txqe
Files Structrue:
| - hw2
| - code
| - Beras
| - eight files extension is .py
| - assignment.py
| - preprocess.py
| - visualize.py
| - data
| - mnist
| - four files of minst dataset
Conceptual Questions
Getting the Stencil
Getting the Data
Assignment Overview
Before Starting
1. Preprocessing the Data
2. One Hot Encoding
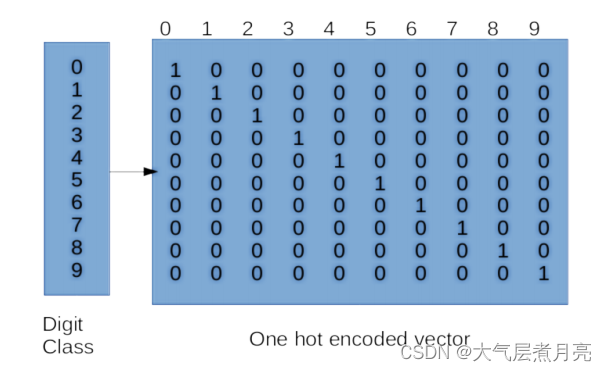
● fit() : [TODO] In this function you need to fetch all the unique labels in thedata (store this in self.uniq ) and create a dictionary with labels as the keysand their corresponding one hot encodings as values. Hint: You might want tolook at np.eye() to get the one-hot encodings. Ultimately, you will store thisdictionary in self.uniq2oh .
● forward() : In this function, we pass a vector of all the actual labels in thetraining set and call fit() to populate the uniq2oh dictionary with uniquelabels and their corresponding one-hot encoding and then use it to return anarray of one-hot encoded labels for each label in the training set. This functionhas already been filled out for you!
● inverse() : In the function, we reverse the one-hot encoding back to the actuallabel. This has already been done for you.For example, if we have labels X and Y with one-hot encodings of [1,0] and [0,1], we’dwant to create a dictionary as follows: {X: [1,0], Y: [0,1]}. As shown in the image above,for MNIST, you will have 10 labels, so your dictionary should have ten entries!You may notice that some classes inherit from Callable or Diffable. More on thisin the next section!
import numpy as np
from .core import Callable
class OneHotEncoder(Callable):
"""
One-Hot Encodes labels. First takes in a candidate set to figure out what elements it
needs to consider, and then one-hot encodes subsequent input datasets in the
forward pass.
SIMPLIFICATIONS:
- Implementation assumes that entries are individual elements.
- Forward will call fit if it hasn't been done yet; most implementations will just error.
- keras does not have OneHotEncoder; has LabelEncoder, CategoricalEncoder, and to_categorical()
"""
def fit(self, data):
"""
Fits the one-hot encoder to a candidate dataset. Said dataset should contain
all encounterable elements.
:param data: 1D array containing labels.
For example, data = [0, 1, 3, 3, 1, 9, ...]
"""
## TODO: Fetch all the unique labels and create a dictionary with
## the unique labels as keys and their one hot encodings as values
## HINT: look up np.eye() and see if you can utilize it!
## HINT: Wouldn't it be nice if we just gave you the implementation somewhere...
self.uniq = None # all the unique labels from `data`
self.uniq2oh = None # a lookup dictionary with labels and corresponding encodings
def forward(self, data):
if not hasattr(self, "uniq2oh"):
self.fit(data)
return np.array([self.uniq2oh[x] for x in data])
def inverse(self, data):
assert hasattr(self, "uniq"), \
"forward() or fit() must be called before attempting to invert"
return np.array([self.uniq[x == 1][0] for x in data])
3. Core Abstractions
● CategoricalAccuracy (./metrics.py): Computes the accuracy of predictedprobabilities against a list of ground-truth labels. As accuracy is not optimized for,there is no need to compute its gradient. Furthermore, categorical accuracy ispiecewise discontinuous, so the gradient would technically be 0 or undefined.
● OneHotEncoder (./onehot.py) : You can one-hot encode a class instance into aprobability distribution to optimize for classifying into discrete options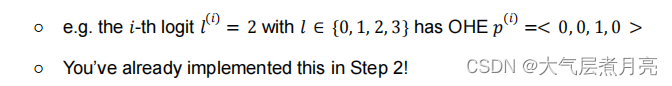
Diffable: A callable which can also be differentiated. We can use these in our pipelineand optimize through them! Thus, most of these classes are made for use in your neuralnetwork layers. These are the ones you’ll need to implement: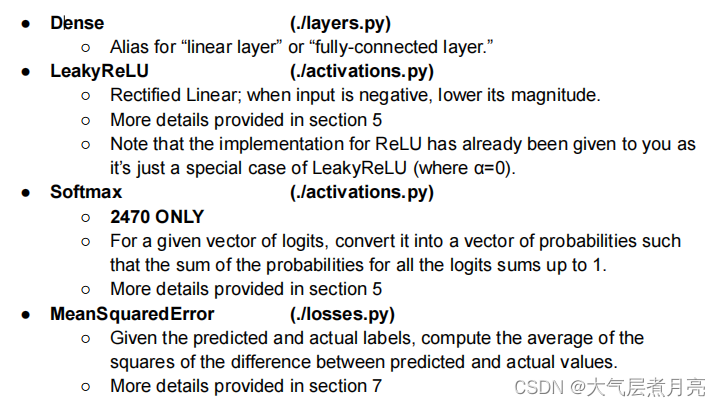
Example: Consider a Dense layer instance. Let s represents the input size (source), drepresents the output size (destination), and b represents the batch size. Then: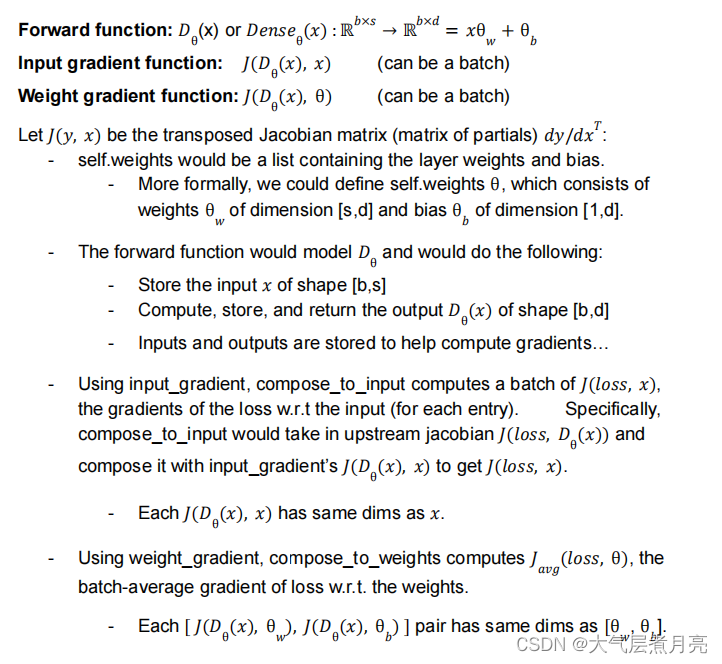
GradientTape: This class will function exactly like tf.GradientTape() (See lab 3).You can think of a GradientTape as a logger. Every time an operation is performed withinthe scope of a GradientTape, it records which operation occurred. Then, duringbackprop, we can compute the gradient for all of the operations. This allows us todifferentiate our final output with respect to any intermediate step. When operations arecomputed outside the scope of GradientTape, they aren’t recorded, so your code willhave no record of them and cannot compute the gradients.You can check out how this is implemented in core! Of course, Tensorflow’s gradienttape implementation is a lot more complicated and involves constructing a graph.● [TODO] Implement the gradient method, which returns a list of gradients corresponding to the list of trainable weights in the network. Details in the code.
from abc import ABC, abstractmethod # # For abstract method support
from typing import Tuple
import numpy as np
## DO NOT MODIFY THIS CLASS
class Callable(ABC):
"""
Callable Sub-classes:
- CategoricalAccuracy (./metrics.py) - TODO
- OneHotEncoder (./preprocess.py) - TODO
- Diffable (.) - DONE
"""
def __call__(self, *args, **kwargs) -> np.array:
"""Lets `self()` and `self.forward()` be the same"""
return self.forward(*args, **kwargs)
@abstractmethod
def forward(self, *args, **kwargs) -> np.array:
"""Pass inputs through function. Can store inputs and outputs as instance variables"""
pass
## DO NOT MODIFY THIS CLASS
class Diffable(Callable):
"""
Diffable Sub-classes:
- Dense (./layers.py) - TODO
- LeakyReLU, ReLU (./activations.py) - TODO
- Softmax (./activations.py) - TODO
- MeanSquaredError (./losses.py) - TODO
"""
"""Stores whether the operation being used is inside a gradient tape scope"""
gradient_tape = None ## All-instance-shared variable
def __init__(self):
"""Is the layer trainable"""
super().__init__()
self.trainable = True ## self-only instance variable
def __call__(self, *args, **kwargs) -> np.array:
"""
If there is a gradient tape scope in effect, perform AND RECORD the operation.
Otherwise... just perform the operation and don't let the gradient tape know.
"""
if Diffable.gradient_tape is not None:
Diffable.gradient_tape.operations += [self]
return self.forward(*args, **kwargs)
@abstractmethod
def input_gradients(self: np.array) -> np.array:
"""Returns gradient for input (this part gets specified for all diffables)"""
pass
def weight_gradients(self: np.array) -> Tuple[np.array, np.array]:
"""Returns gradient for weights (this part gets specified for SOME diffables)"""
return ()
def compose_to_input(self, J: np.array) -> np.array:
"""
Compose the inputted cumulative jacobian with the input jacobian for the layer.
Implemented with batch-level vectorization.
Requires `input_gradients` to provide either batched or overall jacobian.
Assumes input/cumulative jacobians are matrix multiplied
"""
# print(f"Composing to input in {self.__class__.__name__}")
ig = self.input_gradients()
batch_size = J.shape[0]
n_out, n_in = ig.shape[-2:]
j_new = np.zeros((batch_size, n_out), dtype=ig.dtype)
for b in range(batch_size):
ig_b = ig[b] if len(ig.shape) == 3 else ig
j_new[b] = ig_b @ J[b]
return j_new
def compose_to_weight(self, J: np.array) -> list:
"""
Compose the inputted cumulative jacobian with the weight jacobian for the layer.
Implemented with batch-level vectorization.
Requires `weight_gradients` to provide either batched or overall jacobian.
Assumes weight/cumulative jacobians are element-wise multiplied (w/ broadcasting)
and the resulting per-batch statistics are averaged together for avg per-param gradient.
"""
# print(f'Composing to weight in {self.__class__.__name__}')
assert hasattr(
self, "weights"
), f"Layer {self.__class__.__name__} cannot compose along weight path"
J_out = []
## For every weight/weight-gradient pair...
for w, wg in zip(self.weights, self.weight_gradients()):
batch_size = J.shape[0]
## Make a cumulative jacobian which will contribute to the final jacobian
j_new = np.zeros((batch_size, *w.shape), dtype=wg.dtype)
## For every element in the batch (for a single batch-level gradient updates)
for b in range(batch_size):
## If the weight gradient is a batch of transform matrices, get the right entry.
## Allows gradient methods to give either batched or non-batched matrices
wg_b = wg[b] if len(wg.shape) == 3 else wg
## Update the batch's Jacobian update contribution
j_new[b] = wg_b * J[b]
## The final jacobian for this weight is the average gradient update for the batch
J_out += [np.mean(j_new, axis=0)]
## After new jacobian is computed for each weight set, return the list of gradient updatates
return J_out
class GradientTape:
def __init__(self):
## Log of operations that were performed inside tape scope
self.operations = []
def __enter__(self):
# When tape scope is entered, let Diffable start recording to self.operation
Diffable.gradient_tape = self
return self
def __exit__(self, exc_type, exc_val, exc_tb):
# When tape scope is exited, stop letting Diffable record
Diffable.gradient_tape = None
def gradient(self) -> list:
"""Get the gradient from first to last recorded operation"""
## TODO:
##
## Compute weight gradients for all operations.
## If the model has trainable weights [w1, b1, w2, b2] and ends at a loss L.
## the model should return: [dL/dw1, dL/db1, dL/dw2, dL/db2]
##
## Recall that self.operations is populated by Diffable class instances...
##
## Start from the last operation and compute jacobian w.r.t input.
## Continue to propagate the cumulative jacobian through the layer inputs
## until all operations have been differentiated through.
##
## If an operation that has weights is encountered along the way,
## compute the weight gradients and add them to the return list.
## Remember to check if the layer is trainable before doing this though...
grads = []
return grads
4. Layers
● forward() : [TODO] Implement the forward pass and return the outputs.
● weight_gradients() : [TODO] Calculate the gradients with respect to theweights and the biases. This will be used to optimize the layer .
● input_gradients() : [TODO] Calculate the gradients with respect to thelayer inputs. This will be used to propagate the gradient to previous layers.
● _initialize_weight() : [TODO] Initialize the dense layer’s weight values.By default, initialize all the weights to zero (usually a bad idea, by the way). Youare also required to allow for more sophisticated options (when the initializer isset to normal, xavier, and kaiming). Follow Keras math assumptions!○ Normal: Pretty self-explanatory, a unit normal distribution.○ Xavier Normal: Based on keras.GlorotNormal .○ Kaiming He Normal: Based on Keras.HeNormal .You may find np.random.normal helpful while implementing these. The TODOsprovide some justification for why these different initialization methods are necessary butfor more detail, check out this website ! Feel free to add more initializer options!
import numpy as np
from .core import Diffable
class Dense(Diffable):
# https://towardsdatascience.com/weight-initialization-in-neural-networks-a-journey-from-the-basics-to-kaiming-954fb9b47c79
def __init__(self, input_size, output_size, initializer="kaiming"):
super().__init__()
self.w, self.b = self.__class__._initialize_weight(
initializer, input_size, output_size
)
self.weights = [self.w, self.b]
self.inputs = None
self.outputs = None
def forward(self, inputs):
"""Forward pass for a dense layer! Refer to lecture slides for how this is computed."""
self.inputs = inputs
# TODO: implement the forward pass and return the outputs
self.outputs = None
return self.outputs
def weight_gradients(self):
"""Calculating the gradients wrt weights and biases!"""
# TODO: Implement calculation of gradients
wgrads = None
bgrads = None
return wgrads, bgrads
def input_gradients(self):
"""Calculating the gradients wrt inputs!"""
# TODO: Implement calculation of gradients
return None
@staticmethod
def _initialize_weight(initializer, input_size, output_size):
"""
Initializes the values of the weights and biases. The bias weights should always start at zero.
However, the weights should follow the given distribution defined by the initializer parameter
(zero, normal, xavier, or kaiming). You can do this with an if statement
cycling through each option!
Details on each weight initialization option:
- Zero: Weights and biases contain only 0's. Generally a bad idea since the gradient update
will be the same for each weight so all weights will have the same values.
- Normal: Weights are initialized according to a normal distribution.
- Xavier: Goal is to initialize the weights so that the variance of the activations are the
same across every layer. This helps to prevent exploding or vanishing gradients. Typically
works better for layers with tanh or sigmoid activation.
- Kaiming: Similar purpose as Xavier initialization. Typically works better for layers
with ReLU activation.
"""
initializer = initializer.lower()
assert initializer in (
"zero",
"normal",
"xavier",
"kaiming",
), f"Unknown dense weight initialization strategy '{initializer}' requested"
io_size = (input_size, output_size)
# TODO: Implement default assumption: zero-init for weights and bias
# TODO: Implement remaining options (normal, xavier, kaiming initializations). Note that
# strings must be exactly as written in the assert above
return None, None
5. Activation Functions
● LeakyReLU()○ forward() : [TODO] Given input x , compute & return LeakyReLU(x) .○ input_gradients() : [TODO] Compute & return the partial withrespect to inputs by differentiating LeakyReLU .
● Softmax() : (2470 ONLY)forward(): [TODO] Given input x , compute & return Softmax(x) .Make sure you use stable softmax where you subtract max of all entriesto prevent overflow/undvim erflow issues.○ input_gradients() : [TODO] Partial w.r.t. inputs of Softmax() .
import numpy as np
from .core import Diffable
class LeakyReLU(Diffable):
def __init__(self, alpha=0.3):
super().__init__()
self.alpha = alpha
self.inputs = None
self.outputs = None
def forward(self, inputs):
# TODO: Given an input array `x`, compute LeakyReLU(x)
self.inputs = inputs
# Your code here:
self.outputs = None
return self.outputs
def input_gradients(self):
# TODO: Compute and return the gradients
return 0
def compose_to_input(self, J):
# TODO: Maybe you'll want to override the default?
return super().compose_to_input(J)
class ReLU(LeakyReLU):
def __init__(self):
super().__init__(alpha=0)
class Softmax(Diffable):
def __init__(self):
super().__init__()
self.inputs = None
self.outputs = None
def forward(self, inputs):
"""Softmax forward pass!"""
# TODO: Implement
# HINT: Use stable softmax, which subtracts maximum from
# all entries to prevent overflow/underflow issues
self.inputs = inputs
# Your code here:
self.outputs = None
return self.outputs
def input_gradients(self):
"""Softmax backprop!"""
# TODO: Compute and return the gradients
return 0
6. Filling in the model
● compile() : Initialize the model optimizer, loss function, & accuracy function,which are fed in as arguments, for your SequentialModel instance to use.
● fit() : Trains your model to associate input to outputs. Training is repeated foreach epoch, and the data is batched based on argument. It also computesbatch_metrics , epoch_metrics , and the aggregated agg_metrics thatcan be used to track the training progress of your model.
● evaluate() : [TODO] Evaluate the performance of the final model using themetrics mentioned above during the testing phase. It’s almost identical to thefit() function; think about what would change between training and testing).
● call() : [TODO] Hint: what does it mean to call a sequential model? Rememberthat a sequential model is a stack of layers where each layer has exactly oneinput vector and one output vector. You can find this function within theSequentialModel class in assignment.py .
● batch_step() : [TODO] You will observe that fit() calls this function for eachbatch. You will first compute your model predictions for the input batch. In thetraining phase, you will need to compute gradients and update your weightsaccording to the optimizer you are using. For backpropagation during training,you will use GradientTape from the core abstractions ( core.py ) to recordoperations and intermediate values. You will then use the model's optimizer toapply the gradients to your model's trainable variables. Finally, compute andreturn the loss and accuracy for the batch. You can find this function within theSequentialModel class in assignment.py .
from abc import ABC, abstractmethod
from collections import defaultdict
import numpy as np
from .core import Diffable
def print_stats(stat_dict, b=None, b_num=None, e=None, avg=False):
"""
Given a dictionary of names statistics and batch/epoch info,
print them in an appealing manner. If avg, display stat averages.
"""
title_str = " - "
if e is not None:
title_str += f"Epoch {e+1:2}: "
if b is not None:
title_str += f"Batch {b+1:3}"
if b_num is not None:
title_str += f"/{b_num}"
if avg:
title_str += f"Average Stats"
print(f"\r{title_str} : ", end="")
op = np.mean if avg else lambda x: x
print({k: np.round(op(v), 4) for k, v in stat_dict.items()}, end="")
print(" ", end="" if not avg else "\n")
def update_metric_dict(super_dict, sub_dict):
"""
Appends the average of the sub_dict metrics to the super_dict's metric list
"""
for k, v in sub_dict.items():
super_dict[k] += [np.mean(v)]
class Model(ABC):
###############################################################################################
## BEGIN GIVEN
def __init__(self, layers):
"""
Initialize all trainable parameters and take layers as inputs
"""
# Initialize all trainable parameters
assert all([issubclass(layer.__class__, Diffable) for layer in layers])
self.layers = layers
self.trainable_variables = []
for layer in layers:
if hasattr(layer, "weights") and layer.trainable:
for weight in layer.weights:
self.trainable_variables += [weight]
def compile(self, optimizer, loss_fn, acc_fn):
"""
"Compile" the model by taking in the optimizers, loss, and accuracy functions.
In more optimized DL implementations, this will have more involved processes
that make the components extremely efficient but very inflexible.
"""
self.optimizer = optimizer
self.compiled_loss = loss_fn
self.compiled_acc = acc_fn
def fit(self, x, y, epochs, batch_size):
"""
Trains the model by iterating over the input dataset and feeding input batches
into the batch_step method with training. At the end, the metrics are returned.
"""
agg_metrics = defaultdict(lambda: [])
batch_num = x.shape[0] // batch_size
for e in range(epochs):
epoch_metrics = defaultdict(lambda: [])
for b, b1 in enumerate(range(batch_size, x.shape[0] + 1, batch_size)):
b0 = b1 - batch_size
batch_metrics = self.batch_step(x[b0:b1], y[b0:b1], training=True)
update_metric_dict(epoch_metrics, batch_metrics)
print_stats(batch_metrics, b, batch_num, e)
update_metric_dict(agg_metrics, epoch_metrics)
print_stats(epoch_metrics, e=e, avg=True)
return agg_metrics
def evaluate(self, x, y, batch_size):
"""
X is the dataset inputs, Y is the dataset labels.
Evaluates the model by iterating over the input dataset in batches and feeding input batches
into the batch_step method. At the end, the metrics are returned. Should be called on
the testing set to evaluate accuracy of the model using the metrics output from the fit method.
NOTE: This method is almost identical to fit (think about how training and testing differ --
the core logic should be the same)
"""
# TODO: Implement evaluate similarly to fit.
agg_metrics = defaultdict(lambda: [])
return agg_metrics
@abstractmethod
def call(self, inputs):
"""You will implement this in the SequentialModel class in assignment.py"""
return
@abstractmethod
def batch_step(self, x, y, training=True):
"""You will implement this in the SequentialModel class in assignment.py"""
return
7. Loss Function
● forward() : [TODO] Write a function that computes and returns the meansquared error given the predicted and actual labels.Hint: What is MSE?Given the predicted and actual labels, MSE is the average of the squares of thedifferences between predicted and actual values.
● input_gradients() : [TODO] Compute and return the gradients. Usedifferentiation to derive the formula for these gradients.
import numpy as np
from .core import Diffable
class MeanSquaredError(Diffable):
def __init__(self):
super().__init__()
self.y_pred = None
self.y_true = None
self.outputs = None
def forward(self, y_pred, y_true):
"""Mean squared error forward pass!"""
# TODO: Compute and return the MSE given predicted and actual labels
self.y_pred = y_pred
self.y_true = y_true
# Your code here:
self.outputs = None
return None
# https://medium.com/analytics-vidhya/derivative-of-log-loss-function-for-logistic-regression-9b832f025c2d
def input_gradients(self):
"""Mean squared error backpropagation!"""
# TODO: Compute and return the gradients
return None
def clip_0_1(x, eps=1e-8):
return np.clip(x, eps, 1-eps)
class CategoricalCrossentropy(Diffable):
"""
GIVEN. Feel free to use categorical cross entropy as your loss function for your final model.
https://medium.com/analytics-vidhya/derivative-of-log-loss-function-for-logistic-regression-9b832f025c2d
"""
def __init__(self):
super().__init__()
self.truths = None
self.inputs = None
self.outputs = None
def forward(self, inputs, truths):
"""Categorical cross entropy forward pass!"""
# print(truth.shape, inputs.shape)
ll_right = truths * np.log(clip_0_1(inputs))
ll_wrong = (1 - truths) * np.log(clip_0_1(1 - inputs))
nll_total = -np.mean(ll_right + ll_wrong, axis=-1)
self.inputs = inputs
self.truths = truths
self.outputs = np.mean(nll_total, axis=0)
return self.outputs
def input_gradients(self):
"""Categorical cross entropy backpropagation!"""
bn, n = self.inputs.shape
grad = np.zeros(shape=(bn, n), dtype=self.inputs.dtype)
for b in range(bn):
inp = self.inputs[b]
tru = self.truths[b]
grad[b] = inp - tru
grad[b] /= clip_0_1(inp - inp ** 2)
grad[b] /= inp.shape[-1]
return grad
8. Optimizers
● BasicOptimizer : [TODO] A simple optimizer strategy as seen in Lab 1.
● RMSProp : [TODO] Root mean squared error propagation.
● Adam : [TODO] A common adaptive motion estimation-based optimizer.
from collections import defaultdict
import numpy as np
## HINT: Lab 2 might be helpful...
class BasicOptimizer:
def __init__(self, learning_rate):
self.learning_rate = learning_rate
def apply_gradients(self, weights, grads):
# TODO: Update the weights using basic stochastic gradient descent
return
class RMSProp:
def __init__(self, learning_rate, beta=0.9, epsilon=1e-6):
self.learning_rate = learning_rate
self.beta = beta
self.epsilon = epsilon
self.v = defaultdict(lambda: 0)
def apply_gradients(self, weights, grads):
# TODO: Implement RMSProp optimization
# Refer to the lab on Optimizers for a better understanding!
return
class Adam:
def __init__(
self, learning_rate, beta_1=0.9, beta_2=0.999, epsilon=1e-7, amsgrad=False
):
self.amsgrad = amsgrad
self.learning_rate = learning_rate
self.beta_1 = beta_1
self.beta_2 = beta_2
self.epsilon = epsilon
self.m = defaultdict(lambda: 0) # First moment zero vector
self.v = defaultdict(lambda: 0) # Second moment zero vector.
# Expected value of first moment vector
self.m_hat = defaultdict(lambda: 0)
# Expected value of second moment vector
self.v_hat = defaultdict(lambda: 0)
self.t = 0 # Time counter
def apply_gradients(self, weights, grads):
# TODO: Implement Adam optimization
# Refer to the lab on Optimizers for a better understanding!
return
9. Accuracy metrics
● forward() : [TODO] Return the categorical accuracy of your model given thepredicted probabilities and true labels. You should be returning the proportion ofpredicted labels equal to the true labels, where the predicted label for an image isthe label corresponding to the highest probability. Refer to the internet or lectureslides for categorical accuracy math!
import numpy as np
from .core import Callable
class CategoricalAccuracy(Callable):
def forward(self, probs, labels):
"""Categorical accuracy forward pass!"""
super().__init__()
# TODO: Compute and return the categorical accuracy of your model given the output probabilities and true labels
return None10. Train and Test
● A simple model in get_simple_model() with at most one Diffable layer (e.g.,Dense - ./layers.py ) and one activation function (look for them in./activation.py ). This one is provided for you by default, though you canchange it if you’d like. The autograder will evaluate the original one though!
● A slightly more complex model in get_advanced_model() with two or moreDiffable layers and two or more activation functions. We recommend using Adamoptimizer for this model with a decently low learning rate.
from types import SimpleNamespace
import Beras
import numpy as np
class SequentialModel(Beras.Model):
"""
Implemented in Beras/model.py
def __init__(self, layers):
def compile(self, optimizer, loss_fn, acc_fn):
def fit(self, x, y, epochs, batch_size):
def evaluate(self, x, y, batch_size): ## <- TODO
"""
def call(self, inputs):
"""
Forward pass in sequential model. It's helpful to note that layers are initialized in Beras.Model, and
you can refer to them with self.layers. You can call a layer by doing var = layer(input).
"""
# TODO: The call function!
return None
def batch_step(self, x, y, training=True):
"""
Computes loss and accuracy for a batch. This step consists of both a forward and backward pass.
If training=false, don't apply gradients to update the model!
Most of this method (forward, loss, applying gradients)
will take place within the scope of Beras.GradientTape()
"""
# TODO: Compute loss and accuracy for a batch.
# If training, then also update the gradients according to the optimizer
return {"loss": None, "acc": None}
def get_simple_model_components():
"""
Returns a simple single-layer model.
"""
## DO NOT CHANGE IN FINAL SUBMISSION
from Beras.activations import Softmax
from Beras.layers import Dense
from Beras.losses import MeanSquaredError
from Beras.metrics import CategoricalAccuracy
from Beras.optimizers import BasicOptimizer
# TODO: create a model and compile it with layers and functions of your choice
model = SequentialModel([Dense(784, 10), Softmax()])
model.compile(
optimizer=BasicOptimizer(0.02),
loss_fn=MeanSquaredError(),
acc_fn=CategoricalAccuracy(),
)
return SimpleNamespace(model=model, epochs=10, batch_size=100)
def get_advanced_model_components():
"""
Returns a multi-layered model with more involved components.
"""
# TODO: create/compile a model with layers and functions of your choice.
# model = SequentialModel()
return SimpleNamespace(model=None, epochs=None, batch_size=None)
if __name__ == "__main__":
"""
Read in MNIST data and initialize/train/test your model.
"""
from Beras.onehot import OneHotEncoder
import preprocess
## Read in MNIST data,
train_inputs, train_labels = preprocess.get_data_MNIST("train", "../data")
test_inputs, test_labels = preprocess.get_data_MNIST("test", "../data")
## TODO: Use the OneHotEncoder class to one hot encode the labels
ohe = lambda x: 0 ## placeholder function: returns zero for a given input
## Get your model to train and test
simple = False
args = get_simple_model_components() if simple else get_advanced_model_components()
model = args.model
## REMINDER: Threshold of accuracy:
## 1470: >85% on testing accuracy from get_simple_model_components
## 2470: >95% on testing accuracy from get_advanced_model_components
# TODO: Fit your model to the training input and the one hot encoded labels
# Remember to pass all the arguments that SequentialModel.fit() requires
# such as number of epochs and the batch size
train_agg_metrics = model.fit(
train_inputs,
ohe(train_labels),
epochs = args.epochs,
batch_size = args.batch_size
)
## Feel free to use the visualize_metrics function to view your accuracy and loss.
## The final accuracy returned during evaluation must be > 80%.
# from visualize import visualize_images, visualize_metrics
# visualize_metrics(train_agg_metrics["loss"], train_agg_metrics["acc"])
# visualize_images(model, train_inputs, ohe(train_labels))
## TODO: Evaluate your model using your testing inputs and one hot encoded labels.
## This is the number you will be using!
test_agg_metrics = model.evaluate(test_inputs, ohe(test_labels), batch_size=100)
print('Testing Performance:', test_agg_metrics)
11. Visualizing Results
CS1470 Students
- Complete and Submit HW2 Conceptual
- Implement Beras per specifications and make a SequentialModel in assignment.py
- Test the model inside of main
- Get test accuracy >=85% on MNIST with default get_simple_model_components .
- Complete the Exploration notebook and export it to a PDF.
- The “HW2 Intro to Beras” notebook is just for your reference.
CS2470 Students
- Implement Softmax activation function (forward pass and input_gradients )
- Get testing accuracy >95% on MNIST model from get_advanced_model_components .
- You will need to specify a multi-layered model, will have to explorehyperparameter options, and may want to add additional features.
- Additional features may include regularization, other weight initializationschemes, aggregation layers, dropout, rate scheduling, or skip connections. Ifyou have other ideas, feel free to ask publicly on Ed and we’ll let you know if theyare also ok.
- When implementing these features, try to mimic the Keras API as much aspossible. This will help significantly with your Exploration notebook.
- Finish 2470 components for Exploration notebook and conceptual questions.
1. git add file1 file2 file3 (or -A)
2. git commit -m “commit message”
3. git push
After committing and pushing your changes to your repo (which you can check online if you're unsure if it worked), you can now just upload the repo to Gradescope! If you’re testing out code on multiple branches, you have the option to pick whichever one you want.
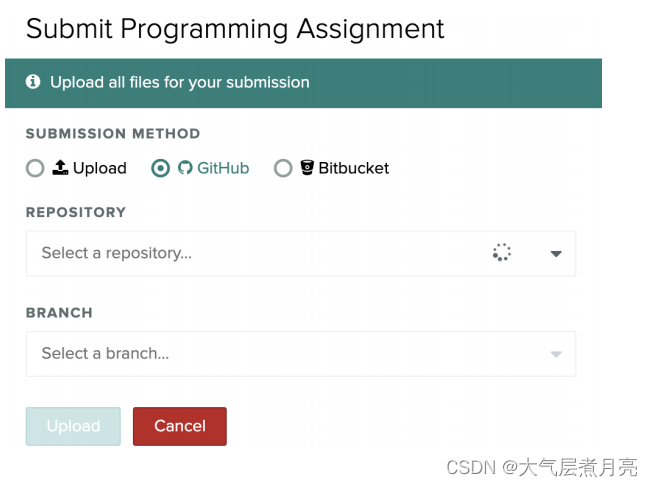
IMPORTANT!
Thanks!

























 4285
4285











 被折叠的 条评论
为什么被折叠?
被折叠的 条评论
为什么被折叠?










