论文阅读笔记(一)——Neural ADMIXTURE for rapid genomic clustering
目录
摘要
随着遗传研究扩展到大规模、日益多样化的生物库,表征大型队列的遗传结构变得越来越重要。常用的方法是将个体基因组分解成分数簇分配,每个簇代表一个DNA变异频率向量。然而,随着生物库规模的迅速增加,这些方法在计算上变得难以处理。在这里,我们提出了Neural ADMIXTURE,这是一种神经网络自编码器,它遵循与当前标准算法ADMIXTURE相同的建模假设,同时减少了计算时间,甚至超过了最快的替代方案。一个月的连续计算使用ADMIXTURE可以减少到只有几个小时的神经ADMIXTURE。multihead方法允许Neural ADMIXTURE在一次运行中计算多个簇数,从而提供进一步的加速。此外,模型可以存储,允许在线性时间内对新数据进行聚类分配,而无需共享训练样本.
简介
PS:
遗传簇(群集) 是指在人群遗传学中通过分析基因组数据将个体或样本根据遗传相似性分类到不同的组中的一个概念。这些簇代表了个体之间共享相似遗传特征的群体,通常与特定的地理、族群或历史背景相关联。
variant(变异) 是指基因组中与参考序列不同的特定点。这些变异可以是单个核苷酸的改变,称为单核苷酸多态性(SNP),也可以是更大片段的插入、删除或复制。基因组的每个变异都可能影响个体的生物学特征,包括疾病易感性、对药物的反应、甚至是外表特征如眼睛颜色。
在遗传学中,特别指“ 下一代测序”(Next Generation Sequencing, NGS) 技术产生的数据集。这种技术与传统的基因测序技术相比,能够以更低的成本和更高的吞吐量对整个基因组进行快速测序。下一代测序技术使得研究人员可以获取大量的遗传数据,这包括但不限于:
- 全基因组测序(Whole Genome Sequencing, WGS):测序个体的全部DNA。
- 外显子测序(Exome Sequencing):只测序编码蛋白的基因部分(外显子)。
- RNA测序(RNA-Seq):分析RNA分子,了解基因的表达情况。
总得来说就是随着人类基因测序数量规模提升,需要预测特征和疾病,但是由于之前的血统样本偏向于欧洲,极端不平衡,需要算法予以纠正。描述遗传数据集群体结构常见方法是每个样本描述为聚类的一组分数,通过无监督方法寻找聚类质心。一般来说,输入variant(变异)是个体的单核苷酸多态性(snp)序列,即基因组中已知在个体之间存在差异的单个位置。还使用了诸如微型卫星等数量较少的小型数据集。人类基因组中有数百万个snp,大多数是双等位基因(两种变体),允许二进制编码。
- 0 用来编码在某个SNP位置上的最常见或参考变异。这意味着,在这个基因位点上,大多数人群中出现频率最高的变异被赋值为0。
- 1 用来编码在同一个SNP位置上的少数或替代变异。这表示这是一个相对较少见的变异,不是大多数人所持有的。
例如,假设在某个特定的SNP位置,大部分人的DNA序列中是碱基"C",而少数人的是碱基"T"。在这种情况下,我们可以将"C"编码为0(参考变异),将"T"编码为1(替代变异)。
这些变异的频率分布在不同的种群之间会因不同的历史而有所不同:创始事件、迁移、隔离和漂移。
论文提出了一个扩展了基因组聚类方法的自编码器:admixture。admixture是作为STRUCTURE8的计算效率高的替代品而开发的,我们现在将这种对效率的追求带到下一代数据集。我们提出的方法Neural admixture遵循与admixture相同的建模假设,但将任务重新构建为基于神经网络的自编码器,在图形和中央图形单元(gpu和cpu)上提供更快的计算时间,同时保持高质量的任务。
模型结构

a、单头架构。输入序列(x)使用线性层(θ1)投影到64维,并由GELU非线性(σ1)处理。通过将64维序列馈送到使用softmax (σ2)激活的k神经元层(θ2参数化)来计算聚类分配估计Q。最后,解码器使用权重为f的线性层输出输入(x)的重建。注意,解码器被限制在这种线性架构中以确保可解释性。
b、H = 3的简单多头例子。64维隐藏向量由不同的权重集(θ2h)独立复制和处理,得到不同维数的向量,对应不同的K值。每个不同的QKh矩阵由不同的解码器矩阵FKh独立处理,产生H个不同的重构。所有参数以端到端方式联合优化。
单头神经ADMIXTURE(Single-head Neural ADMIXTURE)
- 单头架构通常指的是网络中只有一个解码器(decoder),用于处理单一的聚类任务。
- 在单头模式下,神经网络学习一个固定数量的聚类,处理输入数据,并尝试找到最佳的聚类方式来代表数据的基因结构。
- 这种模式适用于聚类任务相对简单或者数据规模较小的情况。
多头神经ADMIXTURE(Multi-head Neural ADMIXTURE)
- 多头架构在同一个神经网络中包含多个解码器,每个解码器负责不同数量的聚类任务。
- 这种架构允许网络在单次训练过程中同时学习并执行多种不同数量的聚类,从而可以快速比较不同聚类数量下的结果,找到最适合数据的聚类数量。
- 多头方法特别适合于需要对聚类数量进行选择或比较的复杂数据集,能够显著提高计算效率和灵活性。
主要区别和应用
- 单头方法在处理较为简单或明确聚类任务时效果良好,计算需求较低。
- 多头方法则在需要探索不同聚类解决方案时更为有效,尤其适用于大规模或多样性数据集,如需要从多个潜在的聚类数量中选择最佳数量时。
Neural ADMIXTURE采用 标准二元交叉熵 进行训练,使其与传统ADMIXTURE模型的目标函数等效(Methods)。两种初始化技术,一种基于主成分分析,另一种基于原型分析,被用作普通网络初始化的替代方案,以加快训练过程并改善结果(补充部分“解码器初始化”)。此外,有两种机制可以通过控制聚类分配的柔软性来整合关于数据集中混合物数量的先验知识:在训练期间应用L2正则化(方法)和softmax回火(补充部分“softmax回火”)。单头和多头方法都可以适用于给定已知训练标签执行常规分类的监督版本(补充部分“监督训练”)。所提出的方法与原始ADMIXTURE框架完全兼容,允许使用ADMIXTURE结果作为神经ADMIXTURE参数的初始化(补充部分“预训练模式”),反之亦然。
Single-head Neural ADMIXTURE
min Q , F L C ( Q , F ) = − ∑ i , j n i j log ( ∑ k q i k f k j ) + ( 2 − n i j ) log ( 1 − ∑ k q i k f k j ) subject to 0 ≤ f k j ≤ 1 ∑ k q i k = 1 q i k ≥ 0 with Q = ( q i k ) and F = ( f k j ) . \begin{array}{ll} \min _{Q, F} \quad & \mathcal{L}_{\mathrm{C}}(Q, F)=-\sum_{i, j} n_{i j} \log \left(\sum_{k} q_{i k} f_{k j}\right)+\left(2-n_{i j}\right) \log \left(1-\sum_{k} q_{i k} f_{k j}\right) \\ \text { subject to } & 0 \leq f_{k j} \leq 1 \\ & \sum_{k} q_{i k}=1 \\ & q_{i k} \geq 0\\ &\text { with } Q=\left(q_{i k}\right) \text { and } F=\left(f_{k j}\right) \text {. } \end{array} minQ,F subject to LC(Q,F)=−∑i,jnijlog(∑kqikfkj)+(2−nij)log(1−∑kqikfkj)0≤fkj≤1∑kqik=1qik≥0 with Q=(qik) and F=(fkj).
设X表示训练样本,其中特征为每个位置的备用等位基因归一化计数,第i个个体的第j个SNP表示为 x i j = n i j 2 ∈ { 0 , 0.5 , 1 } x_{ij} = n_{ij} 2\in \{0,0.5,1\} xij=nij2∈{0,0.5,1}。然后,X≈QF,其中Q为分配,F为每个SNP和群体的备用等位基因频率,式(1)中的负对数似然是X与QF之间的距离。这可以翻译成一个神经网络作为一个自编码器,其中Q = Ψ(X)是由编码器函数Ψ计算的瓶颈,F是解码器权重本身(图1a)。因为Q是在每次前向传递时估计的,而不是作为一个整体来学习训练数据,为了在以前看不见的数据上检索Q分配,我们可以执行一个简单的前向传递,而不是运行优化过程来固定F,与ADMIXTURE不同。
注意,优化问题(式(1))中的限制在体系结构中施加了限制。与 Q = ( q i k ) Q=\left(q_{i k}\right) Q=(qik)和 q i k ≥ 0 q_{i k} \geq 0 qik≥0相关的瓶颈可以通过在编码器输出处应用softmax激活来强制执行,使瓶颈等同于集群分配。
尽管解码器限制(0≤fkj≤1)可以通过对解码器权重应用sigmoid函数来实现,但我们发现,在每个优化步骤之后,将解码器的权重投影到区间[0,1]就足够了,这是投影梯度下降(projected gradient descent)的最常见形式之一。
解码器必须是线性的,不能后跟非线性,因为这会破坏F矩阵的可解释性;解码器权值与聚类质心之间的等价性将会丧失。另一方面,编码器体系结构不受约束,它可以由几个层组成。所提出的体系结构包括一个64维的非线性层,在瓶颈之前具有GELU激活,并直接作用于输入。后者重新缩放数据,使其具有零均值和单位方差。因为每个SNP的平均值是它的频率p,标准差σ是√p(1 - p),所以{0,1}的输入被编码为{-√p 1 - p,√1 - p},从而向网络提供更明确的等位基因频率信息。
ADMIXTURE模型不能像常规自编码器那样精确地重建输入数据,因为输入的SNP基因型序列nij∈{0,1,2}和重建的pij∈[0,1]没有匹配范围。这可以很容易地通过将基因型计数除以2来补救,因此输入数据为 x i j = n i j 2 ∈ { 0 , 0.5 , 1 } x_{ij} = n_{ij} 2\in \{0,0.5,1\} xij=nij2∈{0,0.5,1}。
PS:
- 0 表示在该SNP位置上个体的两个染色体上的等位基因都是最常见的或参考等位基因。例如,如果参考等位基因是"A",那么基因型"AA"会被编码为0。
- 1 表示个体在该SNP位置上的一个染色体上有最常见的等位基因,另一个染色体上有一个不同的、较少见的等位基因(替代等位基因)。例如,如果参考等位基因是"A",而替代等位基因是"G",那么基因型"AG"或"GA"会被编码为1。
- 2 表示个体在该SNP位置上的两个染色体上的等位基因都是替代等位基因。例如,如果替代等位基因是"G",那么基因型"GG"会被编码为2。
此外,我们提出利用编码器权值的Frobenius范数θ上的惩罚项来最小化二进制交叉熵,而不是最小化函数函数(方程(1)):
L N ( Q , F ) = − ∑ i , j x i j log ( ∑ k q i k f k j ) + ( 1 − x i j ) log ( 1 − ∑ k q i k f k j ) + λ ∥ θ ∥ F 2 . \mathcal{L}_{\mathrm{N}}(Q, F)=-\sum_{i, j} x_{i j} \log \left(\sum_{k} q_{i k} f_{k j}\right)+\left(1-x_{i j}\right) \log \left(1-\sum_{k} q_{i k} f_{k j}\right)+\lambda\|\theta\|_{F}^{2} . LN(Q,F)=−i,j∑xijlog(k∑qikfkj)+(1−xij)log(1−k∑qikfkj)+λ∥θ∥F2.
这个正则化项避免了瓶颈中的困难分配,这有助于在训练过程中减少过拟合。
在式(3)中,我们通过式(1)和式(2)证明了所提出的优化问题与ADMIXTURE问题是等价的(不包括正则化项):
L
N
λ
=
0
(
Q
,
F
)
=
−
∑
i
,
j
x
i
j
log
(
∑
k
q
i
k
f
k
j
)
+
(
1
−
x
i
j
)
log
(
1
−
∑
k
q
i
k
f
k
j
)
=
−
∑
i
,
j
n
i
j
2
log
(
∑
k
q
i
k
f
k
j
)
+
(
1
−
n
i
j
2
)
log
(
1
−
∑
k
q
i
k
f
k
j
)
=
=
−
1
2
∑
i
,
j
n
i
j
log
(
∑
k
q
i
k
f
k
j
)
+
(
2
−
n
i
j
)
log
(
1
−
∑
k
q
i
k
f
k
j
)
=
=
1
2
L
C
(
Q
,
F
)
.
\begin{array}{l} \mathcal{L}_{\mathrm{N}}^{\lambda=0}(Q, F)=-\sum_{i, j} x_{i j} \log \left(\sum_{k} q_{i k} f_{k j}\right)+\left(1-x_{i j}\right) \log \left(1-\sum_{k} q_{i k} f_{k j}\right) \\ =-\sum_{i, j} \frac{n_{i j}}{2} \log \left(\sum_{k} q_{i k} f_{k j}\right)+\left(1-\frac{n_{i j}}{2}\right) \log \left(1-\sum_{k} q_{i k} f_{k j}\right)= \\ =-\frac{1}{2} \sum_{i, j} n_{i j} \log \left(\sum_{k} q_{i k} f_{k j}\right)+\left(2-n_{i j}\right) \log \left(1-\sum_{k} q_{i k} f_{k j}\right)= \\ =\frac{1}{2} \mathcal{L}_{\mathrm{C}}(Q, F) . \end{array}
LNλ=0(Q,F)=−∑i,jxijlog(∑kqikfkj)+(1−xij)log(1−∑kqikfkj)=−∑i,j2nijlog(∑kqikfkj)+(1−2nij)log(1−∑kqikfkj)==−21∑i,jnijlog(∑kqikfkj)+(2−nij)log(1−∑kqikfkj)==21LC(Q,F).
Multi-head Neural ADMIXTURE
当然,通过将簇的数量(K)设置为与训练样本的数量或输入的维度(snp的数量)相等,可以获得完美的重构。然而,瓶颈应该理想地捕获关于给定序列的种群结构的基本信息;因此,我们利用了低维瓶颈。
在ADMIXTURE中,必须进行交叉验证以选择种群簇的数量(K),除非已知关于种群祖先数量的特定先验信息。此外,在许多应用程序中,从业者希望观察集群分配如何随着集群数量的增加而变化。随着测序个体和变异数量的增加,由于额外的计算成本,可用于交叉验证的可行簇数迅速减少。作为一种解决方案,Multi-head Neural ADMIXTURE利用编码器计算的64维潜在表示,允许所有簇数同时运行。这个共享表示是针对K, {K1,…,KH}的不同值共同学习的。
图1b显示了在多头体系结构中如何将共享表示拆分为H个不同的头。第i个头由Ki维向量的非线性投影组成,对应于假设数据中有Ki个不同的遗传簇的分配。虽然每个头都可以连接并通过解码器馈送,但这会导致解码器权重F不可解释。因此,每个头部都需要有自己的解码器,因此,检索输入的H个不同的重建。
由于我们有H个重建,我们现在有H个不同的损失值。我们可以通过最小化方程(4)来训练这个架构:
L
M
N
A
(
Q
K
1
,
…
,
H
,
…
,
F
K
1
,
…
,
H
)
=
∑
h
=
1
H
L
N
(
Q
K
h
,
F
K
h
)
,
\mathcal{L}_{\mathrm{MNA}}\left(Q_{K_{1, \ldots, H}, \ldots}, F_{K_{1, \ldots, H}}\right)=\sum_{h=1}^{H} \mathcal{L}_{\mathrm{N}}\left(Q_{K_{h}}, F_{K_{h}}\right),
LMNA(QK1,…,H,…,FK1,…,H)=h=1∑HLN(QKh,FKh),
其中,QKh和FKh分别是第h个头部的聚类分配和每个种群的SNP频率。多头架构允许在一次向前传递中有效地计算H个不同的簇分配,对应于K的H个不同值。然后,从业者可以对结果进行定量和定性分析,以决定哪个K值最适合该数据。
Code
import dask
import dask.array as da
import json
import logging
import math
import numpy as np
import random
import sys
import torch
import torch.nn as nn
import wandb
from pathlib import Path
from tqdm.auto import tqdm
from typing import Iterable, Optional, Tuple, Union
from .modules import NeuralDecoder, NeuralEncoder, ZeroOneClipper
logging.basicConfig(stream=sys.stdout, level=logging.INFO)
log = logging.getLogger(__name__)
class NeuralAdmixture(nn.Module):
"""Instantiate Neural ADMIXTURE model
Args:
ks (Iterable[int]): different numbers of populations to use. If the list contains more than one value, the multi-head version will run.
num_features (int): number of SNPs used to train the network.
encoder_activation (nn.Module, optional): activation function used in the encoder. Defaults to nn.GELU().
P_init (Optional[torch.Tensor], optional): if provided, corresponds to initialization weights as returned by one of the initialization functions. Defaults to None.
lambda_l2 (float, optional): L2 regularization strength in the encoder. Smaller values will give quasi-binary predictions. Defaults to 5e-4.
hidden_size (int, optional): number of neurons in the first linear layer. Defaults to 64.
freeze_decoder (bool, optional): if set to True, the decoder weights are frozen (useful for reusing other results). Defaults to False.
supervised (bool, optional): if set to True, will run in supervised mode. Defaults to False.
supervised_loss_weight (float, optional): weight given to the supervised loss term. Only applied if running in supervised mode. Defaults to 0.05.
"""
def __init__(self, ks: Iterable[int], num_features: int, encoder_activation: nn.Module=nn.GELU(),
P_init: Optional[torch.Tensor]=None, lambda_l2: float=5e-4, hidden_size: int=64,
freeze_decoder: bool=False, supervised: bool=False, supervised_loss_weight: float=0.05) -> None:
super().__init__()
self.ks = ks
self.num_features = num_features
self.hidden_size = hidden_size
self.encoder_activation = encoder_activation
self.supervised = supervised
self.supervised_loss_weight = supervised_loss_weight
self.freeze_decoder = freeze_decoder
self.batch_norm = nn.BatchNorm1d(self.num_features)
self.lambda_l2 = lambda_l2 if lambda_l2 > 1e-8 else 0
self.softmax = nn.Softmax(dim=1)
self.common_encoder = nn.Sequential(
nn.Linear(self.num_features, self.hidden_size, bias=True),
self.encoder_activation,
)
self.multihead_encoder = NeuralEncoder(self.hidden_size, self.ks)
self.decoders = NeuralDecoder(self.ks, num_features, bias=False, inits=P_init, freeze=self.freeze_decoder)
self.clipper = ZeroOneClipper()
self.decoders.decoders.apply(self.clipper)
def forward(self, X: torch.Tensor, only_assignments: bool=False,
only_hidden_states: bool=False) -> Union[torch.Tensor, Iterable[torch.Tensor], Tuple[torch.Tensor, torch.Tensor]]:
"""Forward pass of the model
Args:
X (torch.Tensor): _description_
only_assignments (bool, optional): if set to True, only return the Q values (used in inference mode for speeding up). Defaults to False.
only_hidden_states (bool, optional): if set to True, only return logits of the Q values. Defaults to False.
Returns:
Union[torch.Tensor, Iterable[torch.Tensor], Tuple[torch.Tensor, torch.Tensor]]: SNP reconstruction and Q values (if last two parameters are set to False), otherwise only Q values or logits of Q values.
"""
X = self.batch_norm(X)
enc = self.common_encoder(X)
del X
hid_states = self.multihead_encoder(enc)
if only_hidden_states:
return hid_states
probs = [self.softmax(h) for h in hid_states]
if only_assignments:
return probs
del enc
return self.decoders(probs), probs
def launch_training(self, trX: da.core.Array, optimizer: torch.optim.Optimizer,
loss_f: torch.nn.modules.loss._Loss, num_epochs: int,
device: torch.device, batch_size: int=0, valX: Optional[da.core.Array]=None,
save_every: int=10, save_path: str='../outputs/model.pt',
trY: Optional[Iterable[str]]=None, valY: Optional[Iterable[str]]=None, seed: int=42,
shuffle: bool=True, log_to_wandb: bool=False, tol: float=1e-5, dry_run: bool=False,
warmup_epochs: int=10, Q_inits: Optional[Iterable[torch.Tensor]]=None) -> int:
"""Launch training pipeline of the model
Args:
trX (da.core.Array): training data matrix
optimizer (torch.optim.Optimizer): loaded optimizer to use
loss_f (torch.nn.modules.loss._Loss): instantiated loss function to use
num_epochs (int): maximum number of epochs. Note that the training might stop earlier if the loss is in a plateau.
device (torch.device): device to use for training
batch_size (int, optional): batch size. If 0, will use all the training data in a single batch. Defaults to 0.
valX (Optional[da.core.Array], optional): _description_. Defaults to None.
save_every (int, optional): save a checkpoint after this number of epochs. Defaults to 10.
save_path (str, optional): output path of the trained model. Defaults to '../outputs/model.pt'.
trY (Optional[Iterable[str]], optional): list of training populations per sample. Defaults to None.
valY (Optional[Iterable[str]], optional): list of validation populations per sample. Defaults to None.
seed (int, optional): seed for RNG. Defaults to 42.
shuffle (bool, optional): whether to shuffle the samples at every epoch. Defaults to True.
log_to_wandb (bool, optional): whether to log training to wandb. Defaults to False.
tol (float, optional): tolerance for early stopping. Training will stop if decrease in loss is smaller than this value. Defaults to 1e-5.
dry_run (bool, optional): whether to run a dry run (no output is written to disk). Defaults to False.
warmup_epochs (int, optional): number of warmup epochs to bring Q to a good initial solution. If 0, no warmup is performed. Defaults to 10.
Q_inits(Optional[Iterable[torch.Tensor]], optional): initial Q values for warmup. Defaults to None.
Returns:
int: number of actual training epochs ran
"""
random.seed(seed)
loss_f_supervised, trY_num, valY_num = None, None, None
tr_losses, val_losses = [], []
log.info(f'Will stop optimization when difference in objective function between two subsequent iterations is < {tol} or after {num_epochs} epochs.')
if self.supervised:
log.info('Going to train on supervised mode.')
assert trY is not None, 'Training ground truth ancestries needed for supervised mode'
ancestry_dict = {anc: idx for idx, anc in enumerate(sorted(np.unique([a for a in trY if a != '-'])))}
assert len(ancestry_dict) == self.ks[0], 'Number of ancestries in training ground truth is not equal to the value of k'
ancestry_dict['-'] = -1
to_idx_mapper = np.vectorize(lambda x: ancestry_dict[x])
trY_num = to_idx_mapper(trY[:])
valY_num = to_idx_mapper(valY[:]) if valY is not None else None
loss_f_supervised = nn.CrossEntropyLoss(reduction='mean')
log.info("Bringing training data into memory...")
trX = trX.compute()
log.info("Running warmup epochs...")
if warmup_epochs > 0:
if Q_inits is None:
log.warning("No Q initialization was provided. Skipping warmup epochs.")
else:
self.decoders.freeze()
loss_f_warmup = nn.BCELoss(reduction='mean')
opt_warmup = torch.optim.AdamW(self.common_encoder.parameters(), lr=1e-5)
for wep in range(warmup_epochs):
_, _ = self._run_warmup_epoch(trX, Q_inits, opt_warmup, loss_f_warmup, batch_size, device, shuffle, epoch_num=wep+1)
if not self.decoders.force_freeze:
self.decoders.unfreeze()
log.info("Training...")
for ep in range(num_epochs):
tr_loss, val_loss = self._run_epoch(trX, optimizer, loss_f, batch_size, valX, device, shuffle, loss_f_supervised, trY_num, valY_num, epoch_num=ep+1)
tr_losses.append(tr_loss)
val_losses.append(val_loss)
assert not math.isnan(tr_loss), 'Training loss is NaN'
if log_to_wandb and val_loss is not None:
wandb.log({"tr_loss": tr_loss, "val_loss": val_loss})
elif log_to_wandb and val_loss is None:
wandb.log({"tr_loss": tr_loss})
tr_diff = tr_losses[-2]-tr_losses[-1] if len(tr_losses) > 1 else 'NaN'
val_diff = val_losses[-2]-val_losses[-1] if val_loss is not None and len(val_losses) > 1 else 'NaN'
log.info(f'[METRICS] EPOCH {ep+1}: mean training loss: {tr_loss}, diff: {tr_diff}')
if val_loss is not None:
log.info(f'[METRICS] EPOCH {ep+1}: mean validation loss: {val_loss}, diff: {val_diff}')
if not dry_run and save_every*ep > 0 and ep % save_every == 0:
torch.save(self.state_dict(), save_path)
if ep > 15 and tol > 0 and self._has_converged(tr_diff, tol):
log.info(f'Convergence criteria met. Stopping fit after {ep+1} epochs...')
return ep+1
log.info(f'Max epochs reached. Stopping fit...')
return ep+1
def _get_encoder_norm(self, p: int=2) -> torch.Tensor:
"""Retrieve the sum of the norm of the encoder parameters (for regularization purposes)
Args:
p (int, optional): norm to retrieve. Defaults to 2.
Returns:
torch.Tensor: sum of norms of the encoder parameters
"""
shared_params = torch.cat([x.view(-1) for x in self.common_encoder.parameters()])
multihead_params = torch.cat([x.view(-1) for x in self.multihead_encoder.parameters()])
return torch.norm(shared_params, p)+torch.norm(multihead_params, p)
def _run_step(self, X: torch.Tensor, optimizer: torch.optim.Optimizer, loss_f: torch.nn.modules.loss._Loss,
loss_f_supervised: Optional[torch.nn.modules.loss._Loss], y: Optional[Iterable[str]],
warmup: Optional[bool]=False, Q_inits: Optional[Iterable[torch.Tensor]]=None) -> float:
"""Run a single optimization step
Args:
X (torch.Tensor): mini-batch of data
optimizer (torch.optim.Optimizer): loaded optimizer to use
loss_f (torch.nn.modules.loss._Loss): instantiated loss function to use
loss_f_supervised (Optional[torch.nn.modules.loss._Loss]): instantiated supervied loss function to use
y (Optional[Iterable[str]], optional): list of training populations per sample. Defaults to None.
warmup (Optional[bool], optional): whether to run a warmup step. Defaults to False.
Q_inits (Optional[Iterable[torch.Tensor]], optional): list of Q initialization matrices. Defaults to None.
Returns:
float: loss value of the mini-batch
"""
optimizer.zero_grad(set_to_none=True)
if warmup:
hid_states = self(X, only_assignments=True)
recs = None
else:
recs, hid_states = self(X)
if Q_inits is None and not warmup: # Regular step
loss = sum((loss_f(rec, X) for rec in recs))
elif Q_inits is not None and warmup: # Warmup step
loss = sum((loss_f(h, Q_init) for h, Q_init in zip(hid_states, Q_inits)))
else:
raise ValueError("Cannot provide Q initialization for a regular step")
if loss_f_supervised is not None: # Currently only implemented for single-head architecture!
mask = y > -1
loss += sum((self.supervised_loss_weight*loss_f_supervised(h[mask], y[mask]) for h in hid_states))
if not warmup and self.lambda_l2 > 0:
loss += self.lambda_l2*self._get_encoder_norm(2)**2
del recs, hid_states
loss.backward()
optimizer.step()
self.decoders.decoders.apply(self.clipper)
return loss.item()
def _validate(self, valX: da.core.Array, loss_f: torch.nn.modules.loss._Loss, batch_size: int, device: torch.device,
loss_f_supervised: Optional[torch.nn.modules.loss._Loss]=None, y: Optional[Iterable[str]]=None) -> float:
"""_summary_
Args:
valX (da.core.Array): validation data matrix
loss_f (torch.nn.modules.loss._Loss): instantiated loss function to use
batch_size (int): batch size. Note that the last batch might be smaller.
device (torch.device): device to use for training
loss_f_supervied (Optional[torch.nn.modules.loss._Loss]): instantiated supervied loss function to use
y (Optional[Iterable[str]], optional): list of training populations per sample. Defaults to None.
Returns:
float: loss value of the validation set
"""
acum_val_loss = 0
with torch.inference_mode():
for X, y_b in self.batch_generator(valX, batch_size, y=y if loss_f_supervised is not None else None):
X = X.to(device)
y_b = y_b.to(device) if y_b is not None else None
recs, hid_states = self(X)
acum_val_loss += sum((loss_f(rec, X).item() for rec in recs))
if loss_f_supervised is not None and y_b is not None:
mask = y_b > -1
acum_val_loss += sum((self.supervised_loss_weight*loss_f_supervised(h[mask], y_b[mask]).item() for h in hid_states))
if self.lambda_l2 > 1e-6:
acum_val_loss += self.lambda_l2*self._get_encoder_norm()**2
return acum_val_loss
def batch_generator(self, X, batch_size=0, shuffle=True, y=None, Q_inits=None):
is_inmem = not isinstance(X, da.core.Array)
idxs = [i for i in range(X.shape[0])]
if shuffle:
random.shuffle(idxs)
if batch_size < 1:
batch_size = X.shape[0]
else:
for i in range(0, X.shape[0], batch_size):
with dask.config.set(**{'array.slicing.split_large_chunks': True}):
to_yield_X, to_yield_y, to_yield_Y_warmup = X[sorted(idxs[i:i+batch_size])], None, None
if not is_inmem:
to_yield_X = to_yield_X.compute()
if y is not None:
to_yield_y = torch.as_tensor(y[sorted(idxs[i:i+batch_size])], dtype=torch.int64)
if Q_inits is not None:
to_yield_Y_warmup = [Q[sorted(idxs[i:i+batch_size])] for Q in Q_inits]
yield torch.as_tensor(to_yield_X, dtype=torch.float32), to_yield_y, to_yield_Y_warmup
def _run_epoch(self, trX, optimizer, loss_f, batch_size, valX,
device, shuffle=True, loss_f_supervised=None,
trY=None, valY=None, epoch_num=0):
tr_loss, val_loss = 0, None
self.train()
total_b = trX.shape[0]//batch_size+1
for X, y, _ in tqdm(self.batch_generator(trX, batch_size, shuffle=shuffle, y=trY if loss_f_supervised is not None else None), desc=f"Epoch {epoch_num}", total=total_b):
step_loss = self._run_step(X.to(device), optimizer, loss_f, loss_f_supervised, y.to(device) if y is not None else None)
tr_loss += step_loss
if valX is not None:
self.eval()
val_loss = self._validate(valX, loss_f, batch_size, device, loss_f_supervised, valY)
return tr_loss/trX.shape[0], val_loss/valX.shape[0]
return tr_loss/trX.shape[0], None
def _run_warmup_epoch(self, trX, Q_inits, optimizer, loss_f, batch_size,
device, shuffle=True, epoch_num=0) -> Tuple[float, Union[float, None]]:
tr_loss = 0
total_b = trX.shape[0]//batch_size+1
self.train()
for X, _, Ys in tqdm(self.batch_generator(trX, batch_size, shuffle=shuffle, Q_inits=Q_inits), desc=f"Warmup epoch {epoch_num}", total=total_b):
step_loss = self._run_step(X.to(device), optimizer, loss_f,
None, None, warmup=True,
Q_inits=[Y.to(device) for Y in Ys])
tr_loss += step_loss
log.info(f"Warmup training loss: {tr_loss/trX.shape[0]}")# !
return tr_loss/trX.shape[0], None
@staticmethod
def _has_converged(diff, tol):
return diff < tol
@staticmethod
def _hudsons_fst(pop1, pop2):
'''
Computes Hudson's Fst given variant frequencies of two populations.
'''
try:
num = np.mean(((pop1-pop2)**2))
den = np.mean(np.multiply(pop1, 1-pop2)+np.multiply(pop2, 1-pop1))+1e-7
return num/den
except Exception as e:
log.error(e)
return np.nan
def display_divergences(self):
for i, k in enumerate(self.ks):
dec = self.decoders.decoders[i].weight.data.cpu().numpy()
header = '\t'.join([f'Pop{p}' for p in range(k-1)])
print(f'\nFst divergences between estimated populations: (K = {k})')
print(f'\t{header}')
print('Pop0')
for j in range(1, k):
print(f'Pop{j}', end='')
pop2 = dec[:,j]
for l in range(j):
pop1 = dec[:,l]
fst = self._hudsons_fst(pop1, pop2)
print(f"\t{fst:0.3f}", end="" if l != j-1 else '\n')
return
def save_config(self, name, save_dir):
config = {
'Ks': self.ks,
'num_snps': self.num_features,
}
with open(Path(save_dir)/f"{name}_config.json", 'w') as fb:
json.dump(config, fb)
log.info('Configuration file saved.')
return
结果
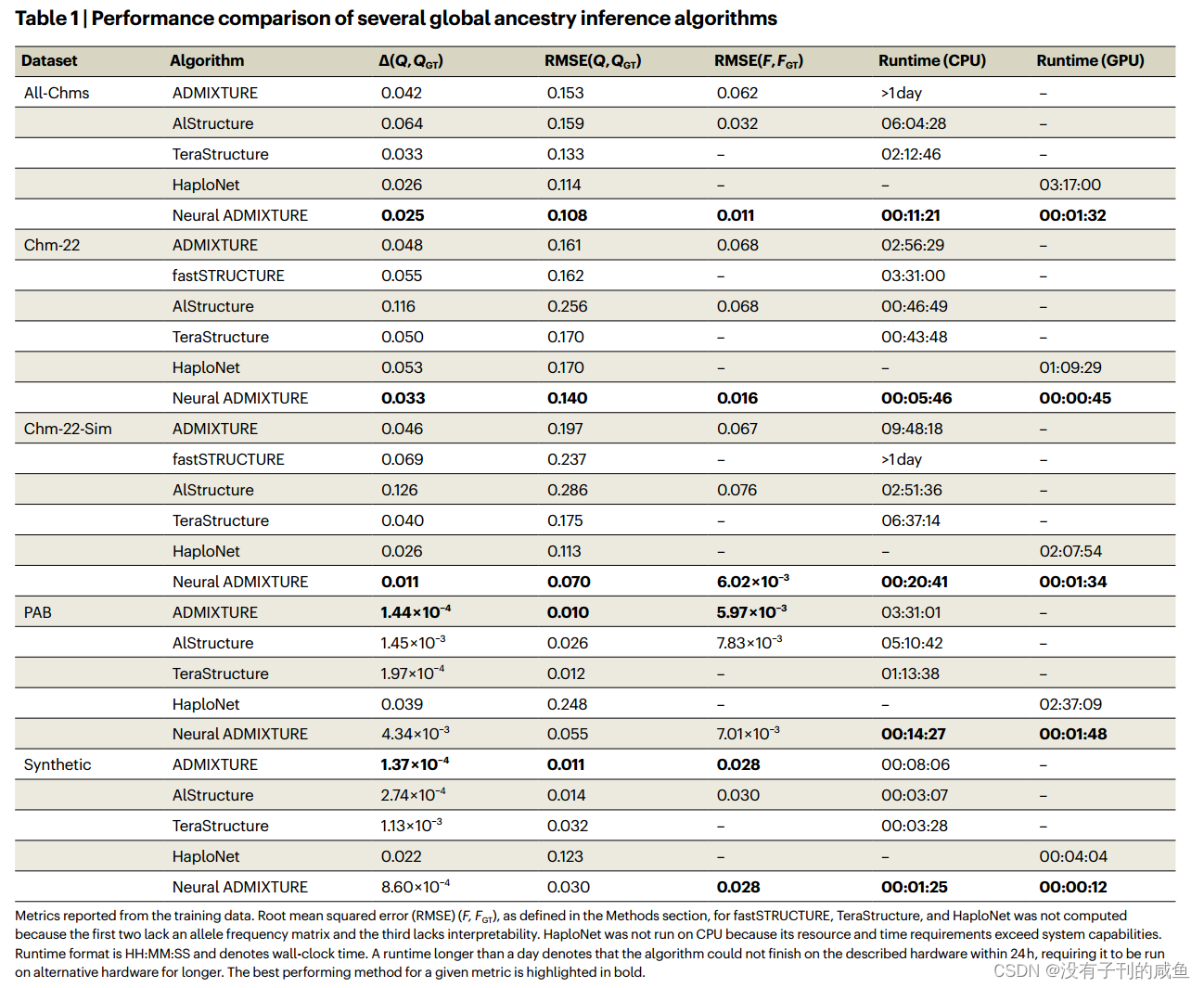
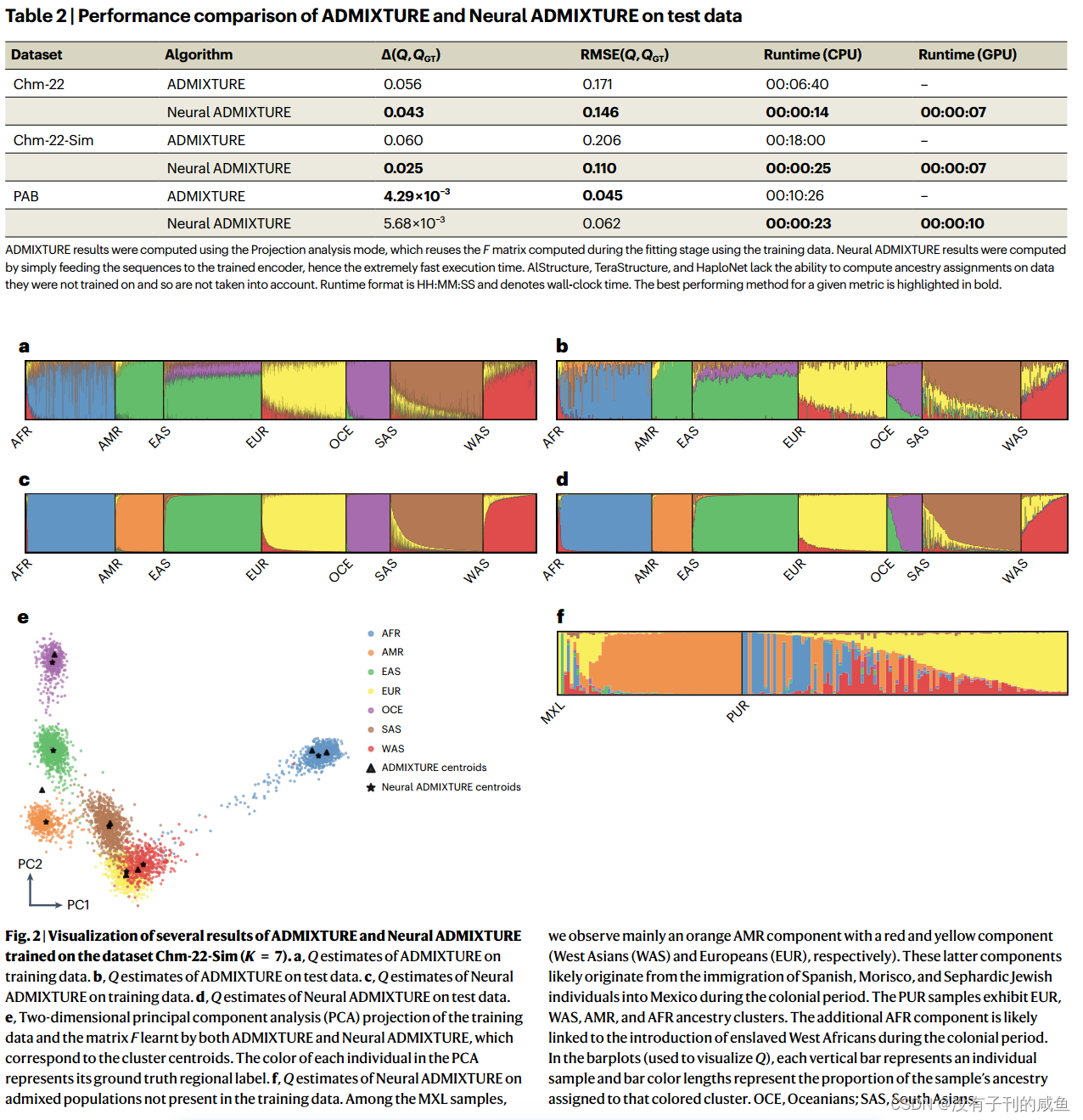
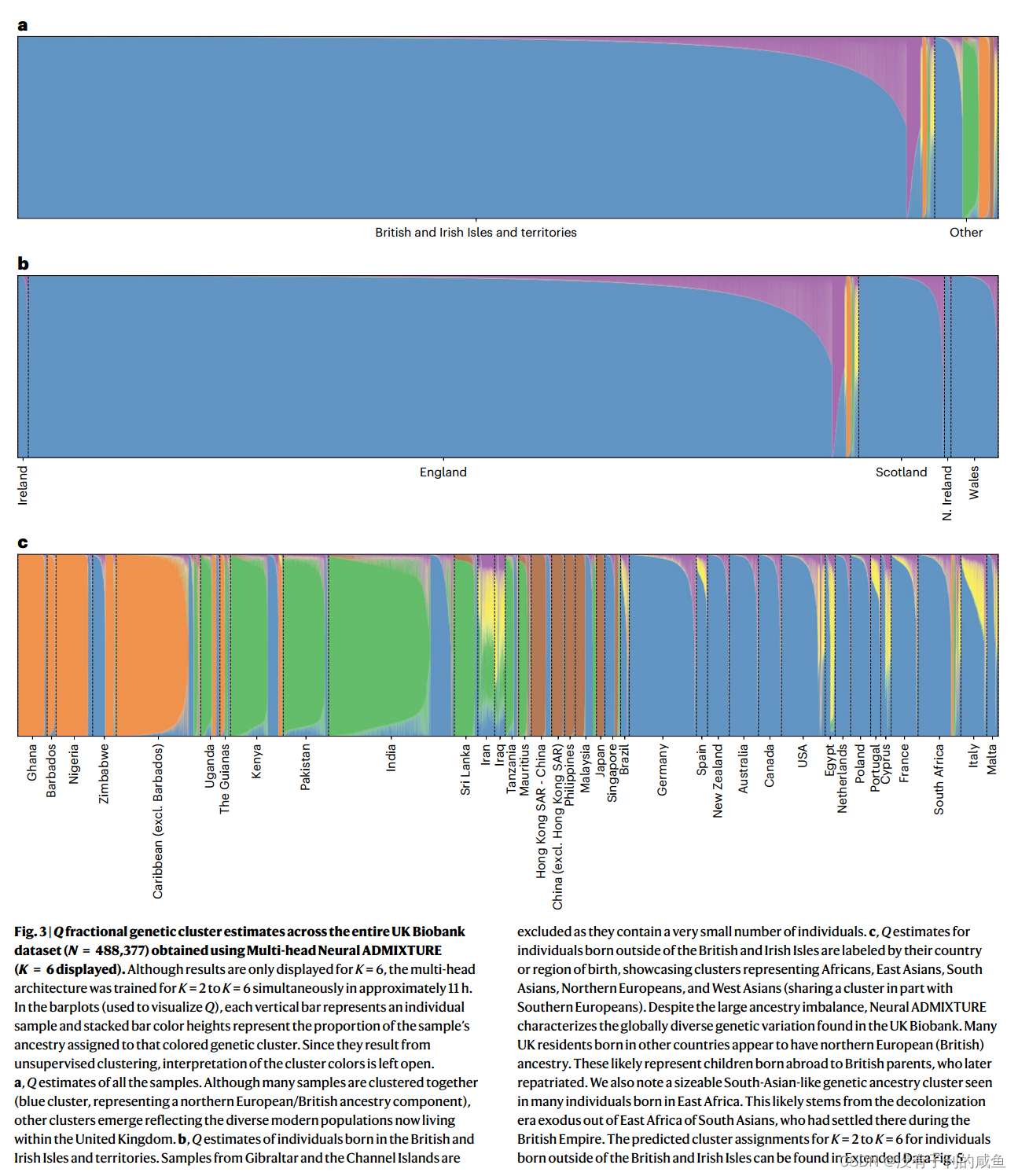
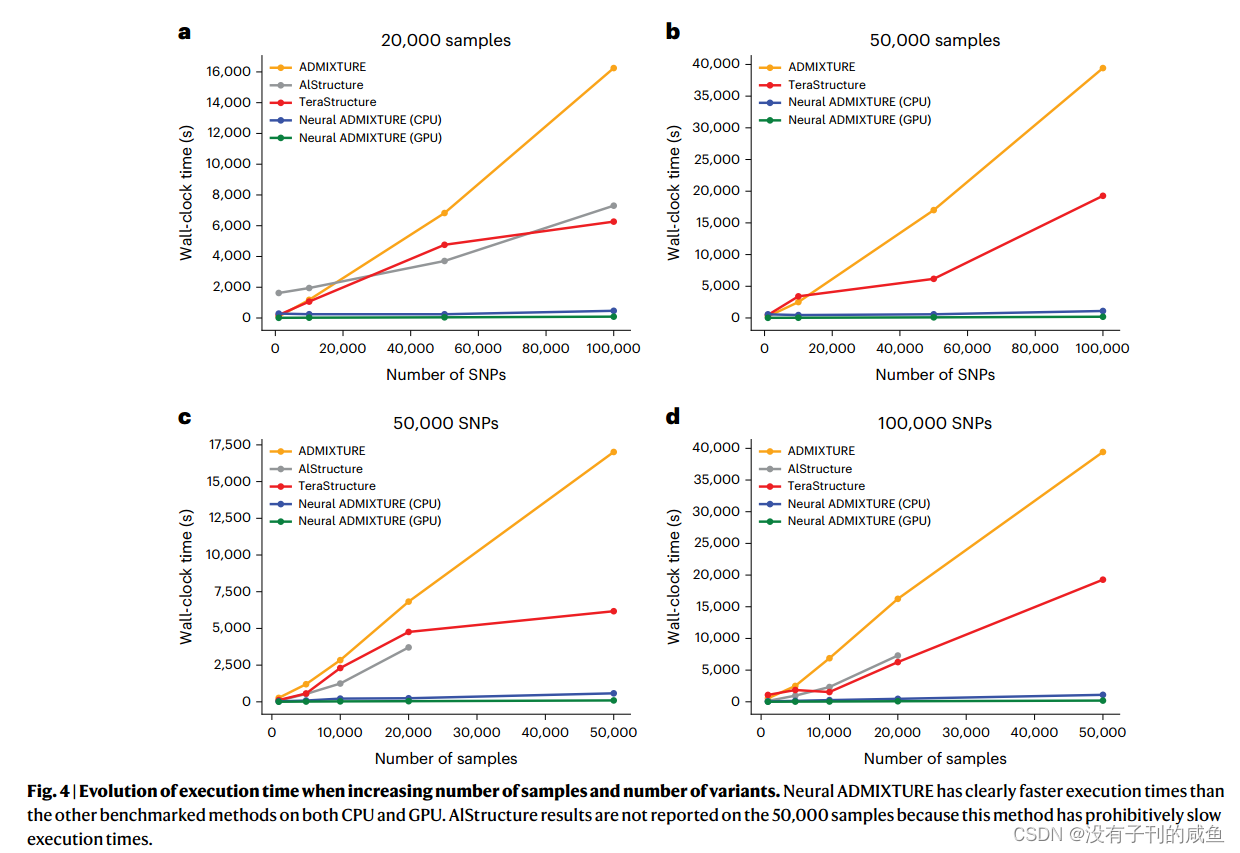




















 717
717











 被折叠的 条评论
为什么被折叠?
被折叠的 条评论
为什么被折叠?








