Gene Ontology(GO)是基因功能国际标准分类体系。GO富集分析是对差异基因等按GO分类,并对分类结果进行基于离散分布的显著性分析、错判率分析、富集度分析,得到与实验目的有显著联系的、低误判率的、靶向性的基因功能分类,该分类即导致样本性状差异的最重要的功能差别。在芯片的数据分析中,研究者可以找出哪些变化基因属于一个共同的GO功能分支,并用统计学方法检定结果是否具有统计学意义,从而得出变化基因主要参与了哪些生物功能。
1. 对mRNA进行显著性功能富集分析,得到具有显著性、靶向性的功能以及显著性功能对应的靶基因。(功能富集)
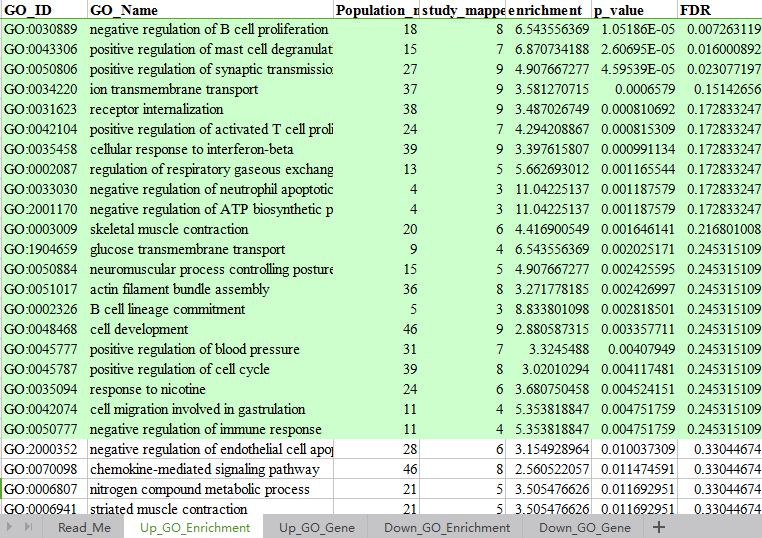
2. 对富集得到的显著性功能以柱状图的形式展示。(功能柱状图)
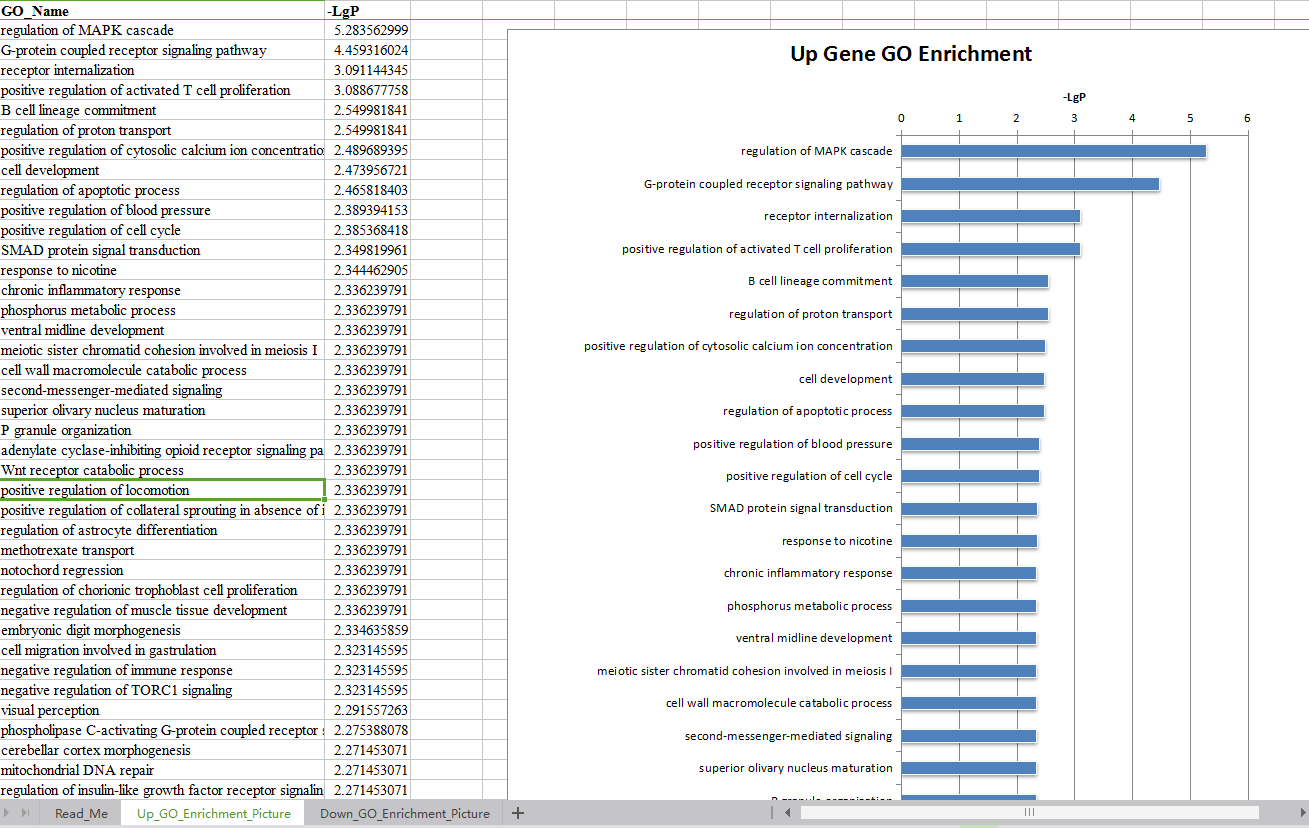
3. 对富集得到的显著性功能以点图的形式展示。(功能点图)





















 9036
9036

 被折叠的 条评论
为什么被折叠?
被折叠的 条评论
为什么被折叠?








