两个阶段:
- 特征提取方法:自回归(Autoregression,AR),公共空间模式(Common Spatial Pattern ,CSP),离散小波变换(Discrete Wavelet Transform,DWT)和功率谱密度( Power Spectral Density,PSD);
- 特征分类方法:Bagging、Boosting、AdaBoost
一、特征提取
共空间模式 Common Spatial Pattern (CSP)
main_CSP
% Extract Common Spatial Pattern (CSP) Feature
close all; clear; clc;
load dataset_BCIcomp1.mat
EEGSignals.x=x_train;
EEGSignals.y=y_train;
Y=y_train;
classLabels = unique(EEGSignals.y);
CSPMatrix = learnCSP(EEGSignals,classLabels);
nbFilterPairs = 1;
X = extractCSP(EEGSignals, CSPMatrix, nbFilterPairs);
EEGSignals.x=x_test;
T = extractCSP(EEGSignals, CSPMatrix, nbFilterPairs);
save dataCSP.mat X Y T
color_L = [0 102 255] ./ 255;
color_R = [255, 0, 102] ./ 255;
pos = find(Y==1);
plot(X(pos,1),X(pos,2),'x','Color',color_L,'LineWidth',2);
hold on
pos = find(Y==2);
plot(X(pos,1),X(pos,2),'o','Color',color_R,'LineWidth',2);
legend('Left Hand','Right Hand')
xlabel('C3','fontweight','bold')
ylabel('C4','fontweight','bold')
函数1:learnCSP
作用:学习CSP生成投影矩阵
function CSPMatrix = learnCSP(EEGSignals,classLabels)
%
%Input:
%EEGSignals: the training EEG signals, composed of 2 classes. These signals
%are a structure such that:
% EEGSignals.x: the EEG signals as a [Ns * Nc * Nt] Matrix where
% Ns: number of EEG samples per trial
% Nc: number of channels (EEG electrodes)
% nT: number of trials
% EEGSignals.y: a [1 * Nt] vector containing the class labels for each trial
% EEGSignals.s: the sampling frequency (in Hz)
%
%Output:
%CSPMatrix: the learnt CSP filters (a [Nc*Nc] matrix with the filters as rows)
%
%See also: extractCSPFeatures
%check and initializations
nbChannels = size(EEGSignals.x,2);
nbTrials = size(EEGSignals.x,3);
nbClasses = length(classLabels);
if nbClasses ~= 2
disp('ERROR! CSP can only be used for two classes');
return;
end
covMatrices = cell(nbClasses,1); %the covariance matrices for each class
%% Computing the normalized covariance matrices for each trial
trialCov = zeros(nbChannels,nbChannels,nbTrials);
for t=1:nbTrials
E = EEGSignals.x(:,:,t)'; %note the transpose
EE = E * E';
trialCov(:,:,t) = EE ./ trace(EE);
end
clear E;
clear EE;
%computing the covariance matrix for each class
for c=1:nbClasses
covMatrices{c} = mean(trialCov(:,:,EEGSignals.y == classLabels(c)),3); %EEGSignals.y==classLabels(c) returns the indeces corresponding to the class labels
end
%the total covariance matrix
covTotal = covMatrices{1} + covMatrices{2};
%whitening transform of total covariance matrix
[Ut Dt] = eig(covTotal); %caution: the eigenvalues are initially in increasing order
eigenvalues = diag(Dt);
[eigenvalues egIndex] = sort(eigenvalues, 'descend');
Ut = Ut(:,egIndex);
P = diag(sqrt(1./eigenvalues)) * Ut';
%transforming covariance matrix of first class using P
transformedCov1 = P * covMatrices{1} * P';
%EVD of the transformed covariance matrix
[U1 D1] = eig(transformedCov1);
eigenvalues = diag(D1);
[eigenvalues egIndex] = sort(eigenvalues, 'descend');
U1 = U1(:, egIndex);
CSPMatrix = U1' * P;
函数2:extractCSP
作用:特征提取
function features = extractCSP(EEGSignals, CSPMatrix, nbFilterPairs)
%
%extract features from an EEG data set using the Common Spatial Patterns (CSP) algorithm
%
%Input:
%EEGSignals: the EEGSignals from which extracting the CSP features. These signals
%are a structure such that:
% EEGSignals.x: the EEG signals as a [Ns * Nc * Nt] Matrix where
% Ns: number of EEG samples per trial
% Nc: number of channels (EEG electrodes)
% nT: number of trials
% EEGSignals.y: a [1 * Nt] vector containing the class labels for each trial
% EEGSignals.s: the sampling frequency (in Hz)
%CSPMatrix: the CSP projection matrix, learnt previously (see function learnCSP)
%nbFilterPairs: number of pairs of CSP filters to be used. The number of
% features extracted will be twice the value of this parameter. The
% filters selected are the one corresponding to the lowest and highest
% eigenvalues
%
%Output:
%features: the features extracted from this EEG data set
% as a [Nt * (nbFilterPairs*2 + 1)] matrix, with the class labels as the
% last column
%
%initializations
nbTrials = size(EEGSignals.x,3);
features = zeros(nbTrials, 2*nbFilterPairs+1);
Filter = CSPMatrix([1:nbFilterPairs (end-nbFilterPairs+1):end],:);
%extracting the CSP features from each trial
for t=1:nbTrials
%projecting the data onto the CSP filters
projectedTrial = Filter * EEGSignals.x(:,:,t)';
%generating the features as the log variance of the projected signals
variances = var(projectedTrial,0,2);
for f=1:length(variances)
features(t,f) = log(1+variances(f));
end
end
原始数据集:3为通道,140为试验trails
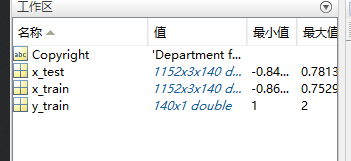
生成的特征数据
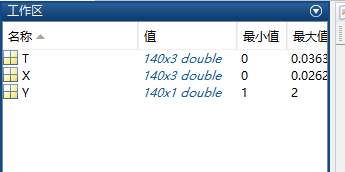
特征散点图:























 3431
3431











 被折叠的 条评论
为什么被折叠?
被折叠的 条评论
为什么被折叠?








