目的:批量生成mCherry+split intein序列,以交给AF-multimer进行结构预测。
所需文件列表:
![]()
文件介绍:
(1)Junction_seqences.dat为连接序列,其格式如下:
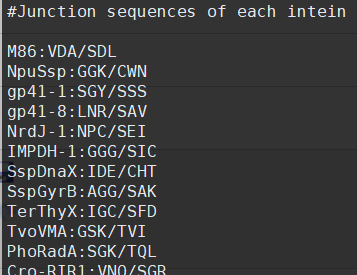
(2)Seq_of_IntN.dat为Intein的N端序列,其格式如下:

(3)Seq_of_IntC.dat为Intein的C端序列,其格式如下:

(4)./seq_for_predict/ 为输出文件存放的文件夹
(5)gene_seq.py脚本的内容如下:
mrc@mrc-Precision-3660:~/project/alphafold2/alphafold2_Vconda/prediction_Split_Int$ cat gene_seq.py
#Python3
#maoruichao@2024.1.8
#Usage: python3 gene_seq.py
##Extein 序列
ExN_seq = 'MVSKGEEDNMAIIKEFMRFKVHMEGSVNGHEFEIEGEGEGRPYEGTQTAKLKVTKGGPLPFAWDILSPQFMYGSKAYVKHPADIPDYLKLSFPEGFKWERVMNFEDGGVVTVTQDSSLQDGEFIYKVKLRGTNFPSDGPVMQKKTMGWEASSERMYPED'
ExC_seq = 'GALKGEIKQRLKLKDGGHYDAEVKTTYKAKKPVQLPGAYNVNIKLDITSHNEDYTIVEQYERAEGRHSTGGMDELYK'
##Open the files
#IntN序列
file_IntN = open('Seq_of_IntN.dat','r')
list_SeqIntN = file_IntN.readlines()
list_SeqIntN_clean = [line.strip() for line in list_SeqIntN if line != '\n' and not line.startswith("#")]
#Linker序列
file_linker = open('Junction_seqences.dat','r')
list_linker = file_linker.readlines()
list_linker_clean = [line.strip() for line in list_linker if line != '\n' and not line.startswith("#")]
#IntC序列
file_IntC = open('Seq_of_IntC.dat','r')
list_SeqIntC = file_IntC.readlines()
list_SeqIntC_clean = [line.strip() for line in list_SeqIntC if line != '\n' and not line.startswith("#")]
#print (len(list_SeqIntN_clean),len(list_SeqIntC_clean))
##For Inteins
#遍历Int_N
num_row = 0
for line_intN in list_SeqIntN_clean:
if line_intN.endswith(':'):
num_row += 1
index1 = line_intN.rfind('-')
if index1 != -1:
int_name = line_intN[:index1]
intN_name = line_intN[:-1]
intN_seq = list_SeqIntN_clean[list_SeqIntN_clean.index(line_intN) + 1]
#获取N-linker序列
for line_linker in list_linker_clean:
list_line = line_linker.split(':')
linker_name = list_line[0]
if int_name == linker_name:
list_linker = list_line[1].split('/')
linker_N = list_linker[0]
#linker_C = list_linker[1]
else:
#print ('int_name != linker_name')
#print ('int_name:', int_name,'\n','linker_name:',linker_name)
pass
#遍历Int_C
num_column = 0
for line_intC in list_SeqIntC_clean:
if line_intC.endswith(':'):
num_column += 1
index2 = line_intC.rfind('-')
if index2 != -1:
int2_name = line_intC[:index2]
intC_name = line_intC[:-1]
intC_seq = list_SeqIntC_clean[list_SeqIntC_clean.index(line_intC) + 1]
#获取C-linker序列
for line_linker in list_linker_clean:
list_line = line_linker.split(':')
linker_name = list_line[0]
if int2_name == linker_name:
list_linker = list_line[1].split('/')
#linker_N = list_linker[0]
linker_C = list_linker[1]
else:
#print ('int_name != linker_name')
#print ('int_name:', int_name,'\n','linker_name:',linker_name)
pass
#生成AF2预测的输入序列
All_seqN = ExN_seq + linker_N + intN_seq
All_seqC = intC_seq + linker_C + ExC_seq
#Output test
print (num_row, num_column, intN_name, intC_name, All_seqN, All_seqC)
#Write to file
filename = './seq_for_predict/' + format(num_row,'02d') + '_' + format(num_column,'02d') + intN_name + '+' + intC_name + '.fasta'
title1 = '>' + intN_name + '|Ex_N+linker+Int_N'
title2 = '>' + intC_name + '|Int_C+linker+Ex_C'
with open(filename, 'w') as newfile:
newfile.write(title1 + '\n')
newfile.write(All_seqN + '\n')
newfile.write(title2 + '\n')
newfile.write(All_seqC + '\n')
else:
pass
else:
pass
#Close file
file_IntN.close()
file_linker.close()
file_IntC.close()
批量预测shell脚本(只能直接运行sh Run_predict.sh,加了nohop后会报错,暂时不清楚原因):
(Alphafold2) mrc@mrc-Precision-3660:speed$ cat ../Run_predict.sh
for filename in `ls *fasta`
do
struc_name="${filename%.*}"
echo "Now start to predict the structure of" $struc_name... >> Run_predict.log
cd /home/mrc/project/alphafold2/alphafold2_Vconda/alphafold-2.3.1
#bash ./run_alphafold.sh -d /home/mrc/project/alphafold2_database -f ../prediction_Split_Int/seq_for_predict/$filename -o ../prediction_Split_Int/seq_for_predict/predict_results/ -t 2023-12-1 -e False -m multimer -l 1
bash ./run_alphafold.sh -d /home/mrc/project/alphafold2_database -f /home/mrc/project/alphafold2/alphafold2_Vconda/prediction_Split_Int/seq_for_predict/$filename -o /home/mrc/project/alphafold2/alphafold2_Vconda/prediction_Split_Int/seq_for_predict/predict_results/ -t 2023-12-1 -e False -m multimer -l 1
cd -
#检查是否预测成功
folder="./predict_results/$struc_name"
genefile_name="ranked_0.pdb"
if [ -e "$folder/$genefile_name" ]; then
echo "The structural prediction for" $struc_name "has been completed" >> Run_predict.log
else
echo "The structural prediction for" $struc_name "is not completed!" >> Run_predict.log
fi
done








 本文介绍了如何通过Python脚本gene_seq.py处理Intein连接序列、N端和C端序列,生成适合AF-multimer结构预测的完整序列,然后在shell脚本Run_predict.sh中调用Alphafold2进行多聚体结构预测。
本文介绍了如何通过Python脚本gene_seq.py处理Intein连接序列、N端和C端序列,生成适合AF-multimer结构预测的完整序列,然后在shell脚本Run_predict.sh中调用Alphafold2进行多聚体结构预测。


















 被折叠的 条评论
为什么被折叠?
被折叠的 条评论
为什么被折叠?








