题目:点击打开链接
题意:
F(1)=a,F(2)=b,F(N)=2F(N-2)+F(N-1)+N^4
给你N,a,b(N,a,b < 2^31)
求F(N)%2147493647
分析:
如果递推式是F(N)=2F(N-2)+F(N-1)就方便了许多,基本的矩阵快速幂
但是多了n^4;
这时,又产生了i^3,i^2,i这些新项需要我们计算
由推出的这些东西 我们就可以构造矩阵了
|1 2 1 4 6 4 1| |f(n+1) | | f(n+2) |
|1 0 0 0 0 0 0| |f(n) | |f(n+1) |
|0 0 1 4 6 4 1| |(n+1)^4| |(n+2)^4|
|0 0 0 1 3 3 1| * |(n+1)^3| = |(n+2)^3|
|0 0 0 0 1 2 1| |(n+1)^2| |(n+2)^2|
|0 0 0 0 0 1 1| | n+1 | | n+2 |
|0 0 0 0 0 0 1| | 1 | | 1 |
#include <bits/stdc++.h>
using namespace std;
typedef long long ll;
const ll mod=2147493647;
int n,a,b;
struct matrix
{
ll m[7][7];
void init1()
{
m[0][0] = b,m[0][1] = 0,m[0][2] = 0,m[0][3] = 0,m[0][4] = 0,m[0][5] = 0,m[0][6] = 0;
m[1][0] = a,m[1][1] = 0,m[1][2] = 0,m[1][3] = 0,m[1][4] = 0,m[1][5] = 0,m[1][6] = 0;
m[2][0] = 16,m[2][1] = 0,m[2][2] = 0,m[2][3] = 0,m[2][4] = 0,m[2][5] = 0,m[2][6] = 0;
m[3][0] = 8,m[3][1] = 0,m[3][2] = 0,m[3][3] = 0,m[3][4] = 0,m[3][5] = 0,m[3][6] = 0;
m[4][0] = 4,m[4][1] = 0,m[4][2] = 0,m[4][3] = 0,m[4][4] = 0,m[4][5] = 0,m[4][6] = 0;
m[5][0] = 2,m[5][1] = 0,m[5][2] = 0,m[5][3] = 0,m[5][4] = 0,m[5][5] = 0,m[5][6] = 0;
m[6][0] = 1,m[6][1] = 0,m[6][2] = 0,m[6][3] = 0,m[6][4] = 0,m[6][5] = 0,m[6][6] = 0;
}
void init2()
{
m[0][0] = 1,m[0][1] = 2,m[0][2] = 1,m[0][3] = 4,m[0][4] = 6,m[0][5] = 4,m[0][6] = 1;
m[1][0] = 1,m[1][1] = 0,m[1][2] = 0,m[1][3] = 0,m[1][4] = 0,m[1][5] = 0,m[1][6] = 0;
m[2][0] = 0,m[2][1] = 0,m[2][2] = 1,m[2][3] = 4,m[2][4] = 6,m[2][5] = 4,m[2][6] = 1;
m[3][0] = 0,m[3][1] = 0,m[3][2] = 0,m[3][3] = 1,m[3][4] = 3,m[3][5] = 3,m[3][6] = 1;
m[4][0] = 0,m[4][1] = 0,m[4][2] = 0,m[4][3] = 0,m[4][4] = 1,m[4][5] = 2,m[4][6] = 1;
m[5][0] = 0,m[5][1] = 0,m[5][2] = 0,m[5][3] = 0,m[5][4] = 0,m[5][5] = 1,m[5][6] = 1;
m[6][0] = 0,m[6][1] = 0,m[6][2] = 0,m[6][3] = 0,m[6][4] = 0,m[6][5] = 0,m[6][6] = 1;
}
};
matrix mul(matrix A,matrix B)
{
matrix ans;
memset(ans.m,0,sizeof(ans.m));
for(int i=0; i<7; i++)
{
for(int j=0; j<7; j++)
{
for(int k=0; k<7; k++)
{
ans.m[i][j]=(ans.m[i][j]+A.m[i][k]*B.m[k][j]%mod)%mod;
}
}
}
return ans;
}
matrix pow(matrix A, ll p)
{
matrix res;
memset(res.m,0,sizeof(res.m));
for(int i=0; i<7; i++)
res.m[i][i]=1;
while(p)
{
//cout<<"___"<<endl;
if(p&1)
res=mul(res,A);
p>>=1;
A=mul(A,A);
}
return res;
}
int main()
{
int T;
scanf("%d",&T);
while(T--)
{
scanf("%d%d%d",&n,&a,&b);
if(n==1)
printf("%lld\n",a%mod);
else if(n==2)
printf("%lld\n",b%mod);
else
{
matrix ans,x;
ans.init1();
x.init2();
x=pow(x,n-1);
ans=mul(x,ans);
printf("%lld\n",ans.m[1][0]);
}
}
return 0;
}










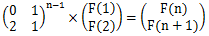
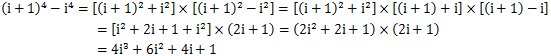

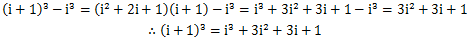
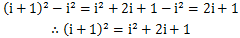
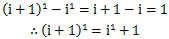














 2913
2913











 被折叠的 条评论
为什么被折叠?
被折叠的 条评论
为什么被折叠?








