VS2013 PCL1.7.2 使用自带eigen库
#include <iostream>
#include <Eigen/Dense>
#include <unsupported/Eigen/NonLinearOptimization>
struct MyFunctor
{
int operator()(const Eigen::VectorXf &x, Eigen::VectorXf &fvec) const
{
double tmp1, tmp2, tmp3;
static const double y[15] = { 1.4e-1, 1.8e-1, 2.2e-1, 2.5e-1, 2.9e-1, 3.2e-1, 3.5e-1,
3.9e-1, 3.7e-1, 5.8e-1, 7.3e-1, 9.6e-1, 1.34, 2.1, 4.39 };
for (int i = 0; i < values(); i++)
{
tmp1 = i + 1;
tmp2 = 16 - i - 1;
tmp3 = (i >= 8) ? tmp2 : tmp1;
fvec[i] = y[i] - (x[0] + tmp1 / (x[1] * tmp2 + x[2] * tmp3));
}
return 0;
}
int df(const Eigen::VectorXf &x, Eigen::MatrixXf &fjac) const
{
double tmp1, tmp2, tmp3, tmp4;
for (int i = 0; i < values(); i++)
{
tmp1 = i + 1;
tmp2 = 16 - i - 1;
tmp3 = (i >= 8) ? tmp2 : tmp1;
tmp4 = (x[1] * tmp2 + x[2] * tmp3); tmp4 = tmp4*tmp4;
fjac(i, 0) = -1;
fjac(i, 1) = tmp1*tmp2 / tmp4;
fjac(i, 2) = tmp1*tmp3 / tmp4;
}
return 0;
}
int inputs() const { return 3; }
int values() const { return 3; } // number of constraints
};
int main(int argc, char *argv[])
{
Eigen::VectorXf x(3);
x(0) = 10;
x(1) = -2;
x(2) = 3;
//x(2) = 0;
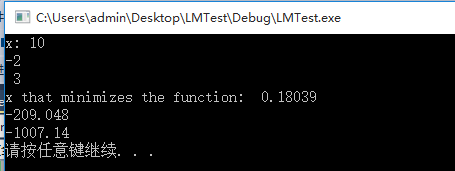
std::cout << "x: " << x << std::endl;
MyFunctor functor;
Eigen::LevenbergMarquardt<MyFunctor, float> lm(functor);
lm.minimize(x);
std::cout << "x that minimizes the function: " << x << std::endl;
system("pause");
return 0;
}结果
sample2:(只参考一下啦)
#include <iostream>
#include <Eigen/Dense>
#include <unsupported/Eigen/NonLinearOptimization>
#include "mex.h"
class pH_Module {
// Functor
private:
double H, H2CO3, HCO3, CO2, CO3, NH3, NH4, HS, H2S, OH;
double Fe, Ca, NO3, SO4, PO4;
double Kc0, Kc1, Kc2, Knh, Khs, Kw;
public:
pH_Module(Eigen::VectorXf x0, Eigen::VectorXf other_conc) {
H = x0(0)*x0(0);
HCO3 = x0(1)*x0(1);
CO2 = x0(2)*x0(2);
CO3 = x0(3)*x0(3);
NH3 = x0(4)*x0(4);
NH4 = x0(5)*x0(5);
HS = x0(6)*x0(6);
H2S = x0(7)*x0(7);
OH = x0(8)*x0(8);
H2CO3 = x0(9)*x0(9);
Fe = other_conc(0)*other_conc(0);
Ca = other_conc(1)*other_conc(1);
NO3 = other_conc(2)*other_conc(2);
SO4 = other_conc(3)*other_conc(3);
PO4 = other_conc(4)*other_conc(4);
Kc0 = 1.7 *pow(10.0, -3.0);
Kc1 = 5.01 *pow(10.0, -7.0) *pow(10.0, 6.0);
Kc2 = 4.78 *pow(10.0, -11.0)*pow(10.0, 6.0);
Knh = 5.62 *pow(10.0, -10.0)*pow(10.0, 6.0);
Khs = 1.3 *pow(10.0, -7.0) *pow(10.0, 6.0);
Kw = pow(10, -14)*pow(10, 12);
};
int operator()(const Eigen::VectorXf &x, Eigen::VectorXf &fvec) const;
int df(const Eigen::VectorXf &x, Eigen::MatrixXf &fjac) const;
int inputs() const;// inputs is the dimension of x.
int values() const; // "values" is the dimension of F
};
int pH_Module::operator()(const Eigen::VectorXf &x, Eigen::VectorXf &fvec) const {
// Equations:
fvec(0) = x(9)*x(9) - Kc0*x(2)*x(2);
fvec(1) = x(0)*x(0)*x(1)*x(1) - Kc1*x(9)*x(9);
fvec(2) = x(0)*x(0)*x(3)*x(3) - Kc2*x(1)*x(1);
fvec(3) = x(0)*x(0)*x(4)*x(4) - Knh*x(5)*x(5);
fvec(4) = x(0)*x(0)*x(6)*x(6) - Khs*x(7)*x(7);
fvec(5) = x(0)*x(0)*x(8)*x(8) - Kw;
fvec(6) = x(4)*x(4) + x(5)*x(5) - NH4 - NH3;
fvec(7) = x(6)*x(6) + x(7)*x(7) - HS - H2S;
fvec(8) = x(3)*x(3) + x(1)*x(1) + x(2)*x(2) + x(9)*x(9) - CO3 - HCO3 - CO2 - H2CO3;
fvec(9) = x(0)*x(0) + x(5)*x(5) + 2 * Fe + 2 * Ca - (x(1)*x(1) + 2 * x(3)*x(3) + x(6)*x(6) + x(8)*x(8) + NO3 + 2 * SO4 + 3 * PO4);
return 0;
}
int pH_Module::df(const Eigen::VectorXf &x, Eigen::MatrixXf &fjac) const {
// Implement Jacobian
// row #0
fjac(0, 0) = 0; fjac(0, 1) = 0; fjac(0, 2) = -2 * Kc0*x(2); fjac(0, 3) = 0; fjac(0, 4) = 0; fjac(0, 5) = 0; fjac(0, 6) = 0; fjac(0, 7) = 0; fjac(0, 8) = 0; fjac(0, 9) = 2 * x(9);
// row #1
fjac(1, 0) = 2 * x(0)*x(1)*x(1); fjac(1, 1) = 2 * x(0)*x(0)*x(1); fjac(1, 2) = 0; fjac(1, 3) = 0; fjac(1, 4) = 0; fjac(1, 5) = 0; fjac(1, 6) = 0; fjac(1, 7) = 0; fjac(1, 8) = 0; fjac(1, 9) = -2 * Kc1*x(9);
// row #2
fjac(2, 0) = 2 * x(0)*x(3)*x(3); fjac(2, 1) = -2 * Kc2*x(1); fjac(2, 2) = 0; fjac(2, 3) = 2 * x(0)*x(0)*x(3); fjac(2, 4) = 0; fjac(2, 5) = 0; fjac(2, 6) = 0; fjac(2, 7) = 0; fjac(2, 8) = 0; fjac(2, 9) = 0;
// row #3
fjac(3, 0) = 2 * x(0)*x(4)*x(4); fjac(3, 1) = 0; fjac(3, 2) = 0; fjac(3, 3) = 0; fjac(3, 4) = 2 * x(0)*x(0)*x(4); fjac(3, 5) = -2 * Knh*x(5); fjac(3, 6) = 0; fjac(3, 7) = 0; fjac(3, 8) = 0; fjac(3, 9) = 0;
// row #4
fjac(4, 0) = 2 * x(0)*x(6)*x(6); fjac(4, 1) = 0; fjac(4, 2) = 0; fjac(4, 3) = 0; fjac(4, 4) = 0; fjac(4, 5) = 0; fjac(4, 6) = 2 * x(0)*x(0)*x(6); fjac(4, 7) = 2 * Khs*x(7); fjac(4, 8) = 0; fjac(4, 9) = 0;
// row #5
fjac(5, 0) = 2 * x(0)*x(8)*x(8); fjac(5, 1) = 0; fjac(5, 2) = 0; fjac(5, 3) = 0; fjac(5, 4) = 0; fjac(5, 5) = 0; fjac(5, 6) = 0; fjac(5, 7) = 0; fjac(5, 8) = 2 * x(0)*x(0)*x(8); fjac(5, 9) = 0;
// row #6
fjac(6, 0) = 0; fjac(6, 1) = 0; fjac(6, 2) = 0; fjac(6, 3) = 0; fjac(6, 4) = 2 * x(4); fjac(6, 5) = 2 * x(5); fjac(6, 6) = 0; fjac(6, 7) = 0; fjac(6, 8) = 0; fjac(6, 9) = 0;
// row #7
fjac(7, 0) = 0; fjac(7, 1) = 0; fjac(7, 2) = 0; fjac(7, 3) = 0; fjac(7, 4) = 0; fjac(7, 5) = 0; fjac(7, 6) = 2 * x(6); fjac(7, 7) = 2 * x(7); fjac(7, 8) = 0; fjac(7, 9) = 0;
// row #8
fjac(8, 0) = 0; fjac(8, 1) = 2 * x(1); fjac(8, 2) = 2 * x(2); fjac(8, 3) = 2 * x(3); fjac(8, 4) = 0; fjac(8, 5) = 0; fjac(8, 6) = 0; fjac(8, 7) = 0; fjac(8, 8) = 0; fjac(8, 9) = 2 * x(9);
// row #9
fjac(9, 0) = 2 * x(0); fjac(9, 1) = -2 * x(1); fjac(9, 2) = 0; fjac(9, 3) = -4 * x(3); fjac(9, 4) = 0; fjac(9, 5) = 2 * x(5); fjac(9, 6) = -2 * x(6); fjac(9, 7) = 0; fjac(9, 8) = -2 * x(8); fjac(9, 9) = 0;
return 0;
}
int pH_Module::inputs() const { return 10; } // inputs is the dimension of x.
int pH_Module::values() const { return 10; } // "values" is the number of f_i
void mexFunction(int nlhs, mxArray *plhs[], int nrhs, const mxArray *prhs[])
{
double *data = (double *)mxGetData(prhs[0]);
Eigen::VectorXf x0(10);
Eigen::VectorXf other_conc(5);
// forming vec of unknowns
for (int i = 0; i < 10; ++i) {
x0(i) = data[i];
}
// vec of other ions
for (int i = 0; i < 5; ++i) {
other_conc(i) = data[i + 10];
}
pH_Module functor(x0, other_conc);
Eigen::LevenbergMarquardt<pH_Module, float> lm(functor);
lm.parameters.xtol = 1e-8;
lm.parameters.ftol = 1e-8;
int info = lm.minimize(x0);
// Assign output
plhs[0] = mxCreateDoubleMatrix(1, 10, mxREAL);
double * z = mxGetPr(plhs[0]);
for (int i = 0; i < 10; ++i)
{
z[i] = x0(i);
}
}





















 679
679

 被折叠的 条评论
为什么被折叠?
被折叠的 条评论
为什么被折叠?








