为什么要介绍医疗模型,因为平时我们工作繁忙,可能身体不舒服会拖着到不得已的时候才到医院,特别是老年人怕麻烦,拖延更严重。如果有了这些模型,我们可以向这些模型提问,给一个初步的了解,同时也可以获取一些养生保健知识。因此这些模型是比较良心,造福人类的。不过如果对于个人医疗需求,请务必咨询合格的医疗保健提供者。
1.医疗大模型介绍
医疗领域的开源 LLM:OpenBioLLM-Llama3,在生物医学领域优于GPT-4、Gemini、Meditron-70B、Med-PaLM-1、Med-PaLM-2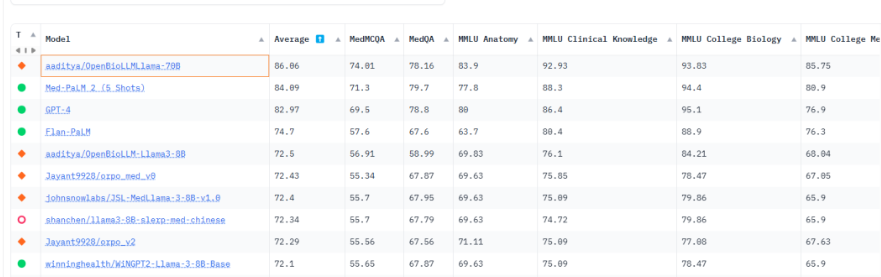
OpenBioLLM-Llama3有两个版本,分别是70B 和 8B
OpenBioLLM-70B提供了SOTA性能,为同等规模模型设立了新的最先进水平
OpenBioLLM-8B模型甚至超越了GPT-3.5、Gemini和Meditron-70B。
- 医疗-LLM排行榜:https://huggingface.co/spaces/openlifescienceai/open_medical_llm_leaderboard
- 70B:https://huggingface.co/aaditya/Llama3-OpenBioLLM-70B
- 8B:https://huggingface.co/aaditya/Llama3-OpenBioLLM-8B
2.安装指南
2.1 下载llama依赖
pip install llama-cpp-python
安装过程
Collecting llama-cpp-python
Downloading llama_cpp_python-0.2.65.tar.gz (38.0 MB)
━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━ 38.0/38.0 MB 42.3 MB/s eta 0:00:00
Installing build dependencies ... done
Getting requirements to build wheel ... done
Installing backend dependencies ... done
Preparing metadata (pyproject.toml) ... done
Requirement already satisfied: typing-extensions>=4.5.0 in /usr/local/lib/python3.10/dist-packages (from llama-cpp-python) (4.11.0)
Requirement already satisfied: numpy>=1.20.0 in /usr/local/lib/python3.10/dist-packages (from llama-cpp-python) (1.25.2)
Collecting diskcache>=5.6.1 (from llama-cpp-python)
Downloading diskcache-5.6.3-py3-none-any.whl (45 kB)
━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━ 45.5/45.5 kB 6.7 MB/s eta 0:00:00
Requirement already satisfied: jinja2>=2.11.3 in /usr/local/lib/python3.10/dist-packages (from llama-cpp-python) (3.1.3)
Requirement already satisfied: MarkupSafe>=2.0 in /usr/local/lib/python3.10/dist-packages (from jinja2>=2.11.3->llama-cpp-python) (2.1.5)
Building wheels for collected packages: llama-cpp-python
Building wheel for llama-cpp-python (pyproject.toml) ... done
Created wheel for llama-cpp-python: filename=llama_cpp_python-0.2.65-cp310-cp310-linux_x86_64.whl size=39397391 sha256=6f91e47e67bea9fd5cae38ebcc05ea19b6c344a1a609a9d497e4e92e026b611a
Stored in directory: /root/.cache/pip/wheels/46/37/bf/f7c65dbafa5b3845795c23b6634863c1fdf0a9f40678de225e
Successfully built llama-cpp-python
Installing collected packages: diskcache, llama-cpp-python
Successfully installed diskcache-5.6.3 llama-cpp-python-0.2.65
2.2 下载模型
from huggingface_hub import hf_hub_download
from llama_cpp import Llama
model_name = "aaditya/OpenBioLLM-Llama3-8B-GGUF"
model_file = "openbiollm-llama3-8b.Q5_K_M.gguf"
model_path = hf_hub_download(model_name,
filename=model_file,
local_dir='/content')
print("My model path: ", model_path)
llm = Llama(model_path=model_path,
n_gpu_layers=-1)
安装过程
openbiollm-llama3-8b.Q5_K_M.gguf: 100%
5.73G/5.73G [00:15<00:00, 347MB/s]
llama_model_loader: loaded meta data with 22 key-value pairs and 291 tensors from /content/openbiollm-llama3-8b.Q5_K_M.gguf (version GGUF V3 (latest))
llama_model_loader: Dumping metadata keys/values. Note: KV overrides do not apply in this output.
llama_model_loader: - kv 0: general.architecture str = llama
llama_model_loader: - kv 1: general.name str = .
llama_model_loader: - kv 2: llama.vocab_size u32 = 128256
llama_model_loader: - kv 3: llama.context_length u32 = 8192
llama_model_loader: - kv 4: llama.embedding_length u32 = 4096
llama_model_loader: - kv 5: llama.block_count u32 = 32
llama_model_loader: - kv 6: llama.feed_forward_length u32 = 14336
llama_model_loader: - kv 7: llama.rope.dimension_count u32 = 128
llama_model_loader: - kv 8: llama.attention.head_count u32 = 32
llama_model_loader: - kv 9: llama.attention.head_count_kv u32 = 8
llama_model_loader: - kv 10: llama.attention.layer_norm_rms_epsilon f32 = 0.000010
llama_model_loader: - kv 11: llama.rope.freq_base f32 = 500000.000000
llama_model_loader: - kv 12: general.file_type u32 = 17
llama_model_loader: - kv 13: tokenizer.ggml.model str = gpt2
llama_model_loader: - kv 14: tokenizer.ggml.tokens arr[str,128256] = ["!", "\"", "#", "$", "%", "&", "'", ...
llama_model_loader: - kv 15: tokenizer.ggml.scores arr[f32,128256] = [0.000000, 0.000000, 0.000000, 0.0000...
llama_model_loader: - kv 16: tokenizer.ggml.token_type arr[i32,128256] = [1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, ...
llama_model_loader: - kv 17: tokenizer.ggml.merges arr[str,280147] = ["Ġ Ġ", "Ġ ĠĠĠ", "ĠĠ ĠĠ", "...
llama_model_loader: - kv 18: tokenizer.ggml.bos_token_id u32 = 128000
llama_model_loader: - kv 19: tokenizer.ggml.eos_token_id u32 = 128001
llama_model_loader: - kv 20: tokenizer.ggml.padding_token_id u32 = 128001
llama_model_loader: - kv 21: general.quantization_version u32 = 2
llama_model_loader: - type f32: 65 tensors
My model path: /content/openbiollm-llama3-8b.Q5_K_M.gguf
llama_model_loader: - type q5_K: 193 tensors
llama_model_loader: - type q6_K: 33 tensors
llm_load_vocab: special tokens definition check successful ( 256/128256 ).
llm_load_print_meta: format = GGUF V3 (latest)
llm_load_print_meta: arch = llama
llm_load_print_meta: vocab type = BPE
llm_load_print_meta: n_vocab = 128256
llm_load_print_meta: n_merges = 280147
llm_load_print_meta: n_ctx_train = 8192
llm_load_print_meta: n_embd = 4096
llm_load_print_meta: n_head = 32
llm_load_print_meta: n_head_kv = 8
llm_load_print_meta: n_layer = 32
llm_load_print_meta: n_rot = 128
llm_load_print_meta: n_embd_head_k = 128
llm_load_print_meta: n_embd_head_v = 128
llm_load_print_meta: n_gqa = 4
llm_load_print_meta: n_embd_k_gqa = 1024
llm_load_print_meta: n_embd_v_gqa = 1024
llm_load_print_meta: f_norm_eps = 0.0e+00
llm_load_print_meta: f_norm_rms_eps = 1.0e-05
llm_load_print_meta: f_clamp_kqv = 0.0e+00
llm_load_print_meta: f_max_alibi_bias = 0.0e+00
llm_load_print_meta: f_logit_scale = 0.0e+00
llm_load_print_meta: n_ff = 14336
llm_load_print_meta: n_expert = 0
llm_load_print_meta: n_expert_used = 0
llm_load_print_meta: causal attn = 1
llm_load_print_meta: pooling type = 0
llm_load_print_meta: rope type = 0
llm_load_print_meta: rope scaling = linear
llm_load_print_meta: freq_base_train = 500000.0
llm_load_print_meta: freq_scale_train = 1
llm_load_print_meta: n_yarn_orig_ctx = 8192
llm_load_print_meta: rope_finetuned = unknown
llm_load_print_meta: ssm_d_conv = 0
llm_load_print_meta: ssm_d_inner = 0
llm_load_print_meta: ssm_d_state = 0
llm_load_print_meta: ssm_dt_rank = 0
llm_load_print_meta: model type = 8B
llm_load_print_meta: model ftype = Q5_K - Medium
llm_load_print_meta: model params = 8.03 B
llm_load_print_meta: model size = 5.33 GiB (5.70 BPW)
llm_load_print_meta: general.name = .
llm_load_print_meta: BOS token = 128000 '<|begin_of_text|>'
llm_load_print_meta: EOS token = 128001 '<|end_of_text|>'
llm_load_print_meta: PAD token = 128001 '<|end_of_text|>'
llm_load_print_meta: LF token = 128 'Ä'
llm_load_print_meta: EOT token = 128009 '<|eot_id|>'
llm_load_tensors: ggml ctx size = 0.30 MiB
llm_load_tensors: offloading 32 repeating layers to GPU
llm_load_tensors: offloading non-repeating layers to GPU
llm_load_tensors: offloaded 33/33 layers to GPU
llm_load_tensors: CPU buffer size = 344.44 MiB
llm_load_tensors: CUDA0 buffer size = 5115.49 MiB
.........................................................................................
llama_new_context_with_model: n_ctx = 512
llama_new_context_with_model: n_batch = 512
llama_new_context_with_model: n_ubatch = 512
llama_new_context_with_model: freq_base = 500000.0
llama_new_context_with_model: freq_scale = 1
llama_kv_cache_init: CUDA0 KV buffer size = 64.00 MiB
llama_new_context_with_model: KV self size = 64.00 MiB, K (f16): 32.00 MiB, V (f16): 32.00 MiB
llama_new_context_with_model: CUDA_Host output buffer size = 0.49 MiB
llama_new_context_with_model: CUDA0 compute buffer size = 258.50 MiB
llama_new_context_with_model: CUDA_Host compute buffer size = 9.01 MiB
llama_new_context_with_model: graph nodes = 1030
llama_new_context_with_model: graph splits = 2
AVX = 1 | AVX_VNNI = 0 | AVX2 = 1 | AVX512 = 1 | AVX512_VBMI = 0 | AVX512_VNNI = 1 | FMA = 1 | NEON = 0 | ARM_FMA = 0 | F16C = 1 | FP16_VA = 0 | WASM_SIMD = 0 | BLAS = 1 | SSE3 = 1 | SSSE3 = 1 | VSX = 0 | MATMUL_INT8 = 0 | LAMMAFILE = 1 |
Model metadata: {'tokenizer.ggml.padding_token_id': '128001', 'tokenizer.ggml.eos_token_id': '128001', 'general.quantization_version': '2', 'tokenizer.ggml.model': 'gpt2', 'general.architecture': 'llama', 'llama.rope.freq_base': '500000.000000', 'llama.context_length': '8192', 'general.name': '.', 'llama.vocab_size': '128256', 'general.file_type': '17', 'llama.embedding_length': '4096', 'llama.feed_forward_length': '14336', 'llama.attention.layer_norm_rms_epsilon': '0.000010', 'llama.rope.dimension_count': '128', 'tokenizer.ggml.bos_token_id': '128000', 'llama.attention.head_count': '32', 'llama.block_count': '32', 'llama.attention.head_count_kv': '8'}
Using fallback chat format: None
2.3 提问问题
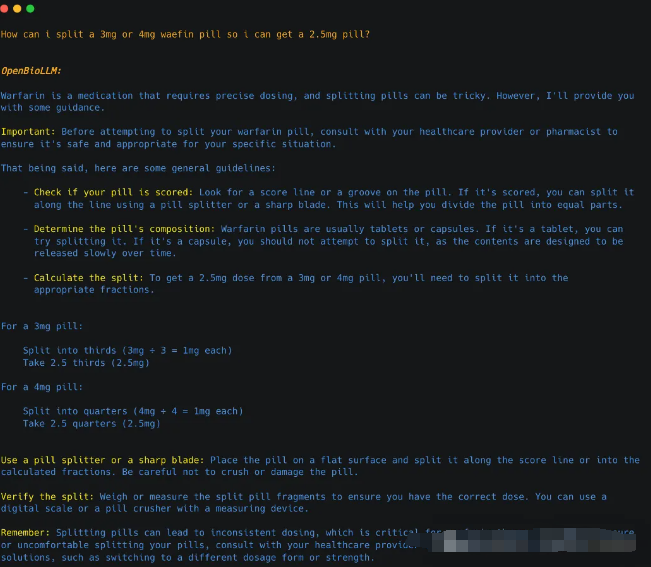
Question = "How can i split a 3mg or 4mg waefin pill so i can get a 2.5mg pill?"
prompt = f"You are an expert and experienced from the healthcare and biomedical domain with extensive medical knowledge and practical experience. Your name is OpenBioLLM, and you were developed by Saama AI Labs with Open Life Science AI. who's willing to help answer the user's query with explanation. In your explanation, leverage your deep medical expertise such as relevant anatomical structures, physiological processes, diagnostic criteria, treatment guidelines, or other pertinent medical concepts. Use precise medical terminology while still aiming to make the explanation clear and accessible to a general audience. Medical Question: {Question} Medical Answer:"
response = llm(prompt, max_tokens=4000)['choices'][0]['text']
print("\n\n\n", response)
结果展示
Llama.generate: prefix-match hit
llama_print_timings: load time = 10599.68 ms
llama_print_timings: sample time = 412.74 ms / 200 runs ( 2.06 ms per token, 484.57 tokens per second)
llama_print_timings: prompt eval time = 0.00 ms / 1 tokens ( 0.00 ms per token, inf tokens per second)
llama_print_timings: eval time = 2192.19 ms / 200 runs ( 10.96 ms per token, 91.23 tokens per second)
llama_print_timings: total time = 4622.41 ms / 201 tokens
To split a 3mg or 4mg Waefin pill into a 2.5mg dose, follow these steps: 1. Use a pill splitter or a sharp knife to divide the pill in half. 2. If using a pill splitter, place the pill in the device and apply even pressure to cut it evenly. 3. If using a knife, carefully place the pill on a non-stick surface and use a sharp blade to slice it into two equal portions. 4. To ensure accuracy, weigh each half-pill on a scale until you find one that weighs approximately 1250mg (which will be close to 2.5mg). 5. Once you have identified the correct half-pill for your desired dosage, consume it as directed by your healthcare provider. It is important to note that pill splitting should only be performed with certain medications under the guidance of a healthcare professional. Always consult with your doctor or pharmacist before attempting to split any medication.
3.用例和示例
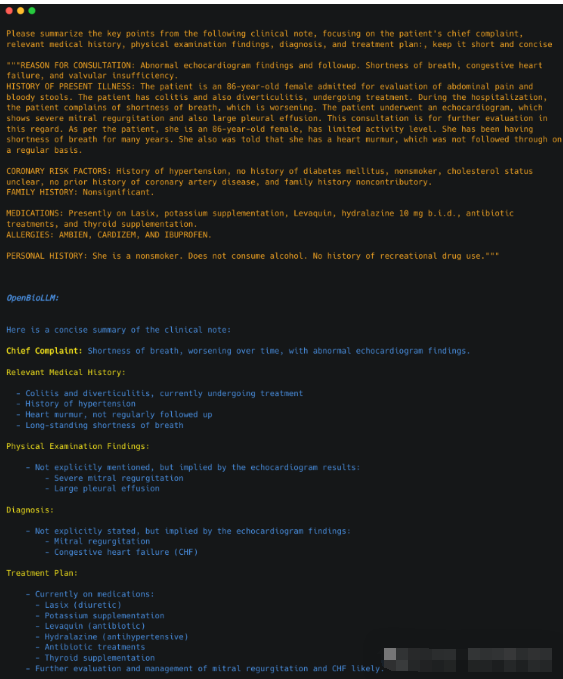
3.1.总结临床笔记
3.2 回答医疗问题
临床实体识别OpenBioLLM-70B可以通过从非结构化临床文本中识别和提取关键的医学概念,如疾病、症状、药物、程序和解剖结构,进行先进的临床实体识别。通过利用其对医学术语和上下文的深刻理解,该模型可以准确地对临床实体进行注释和分类,从而从电子健康记录、研究文章和其他生物医学文本源中实现更高效的信息检索、数据分析和知识发现。此功能可以支持各种下游应用,例如临床决策支持、药物警戒和医学研究。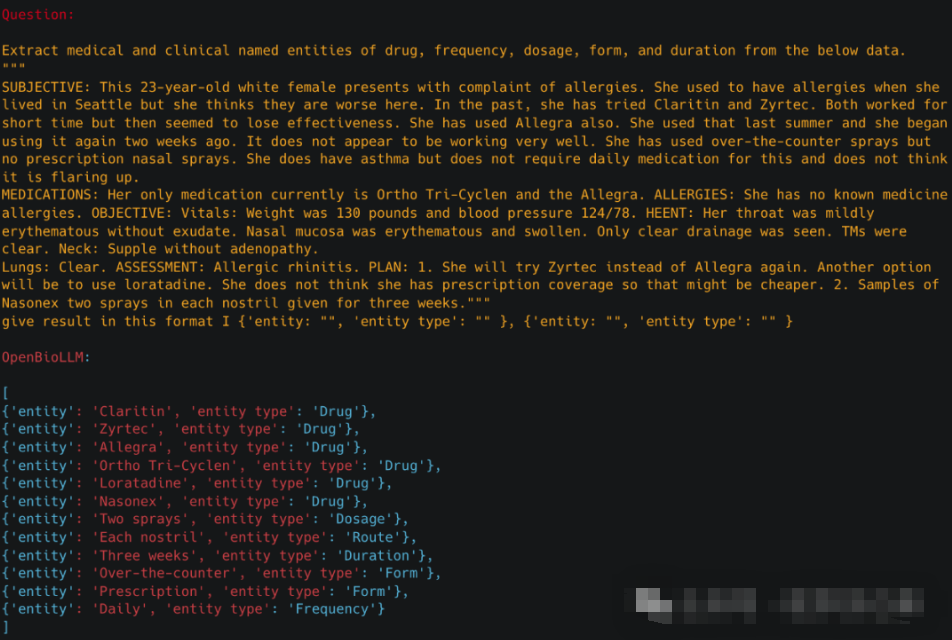
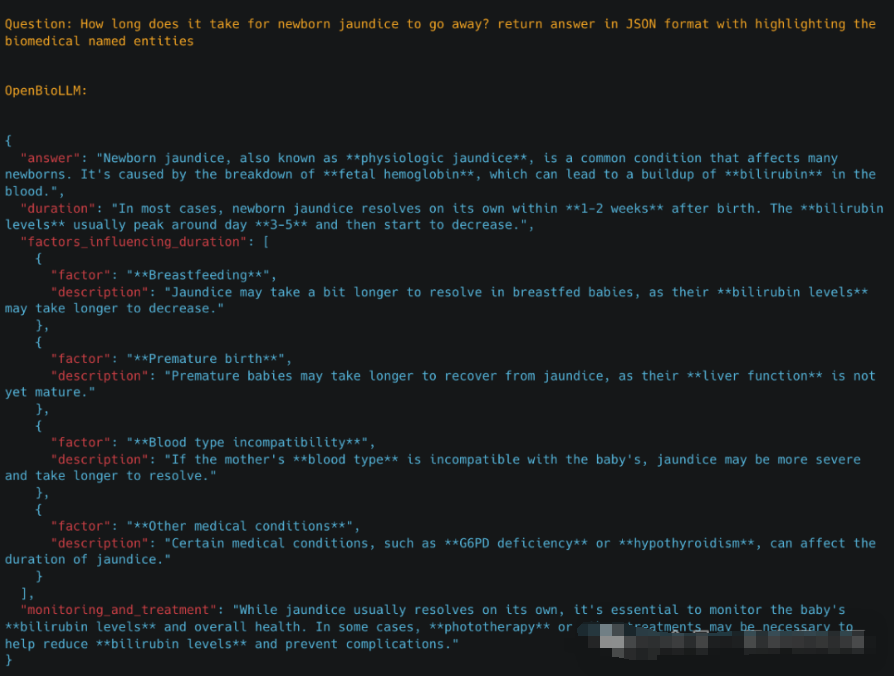
*3.3 生物标志物提取*
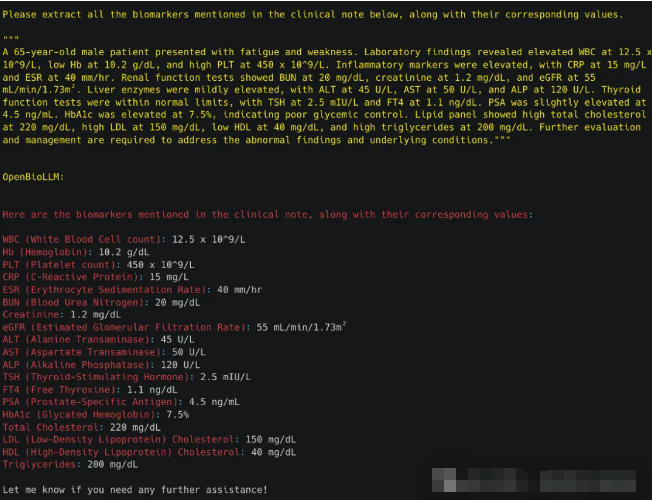
3.4 分类
OpenBioLLM-70B可以执行各种生物医学分类任务,如疾病预测、情感分析、医疗文档分类等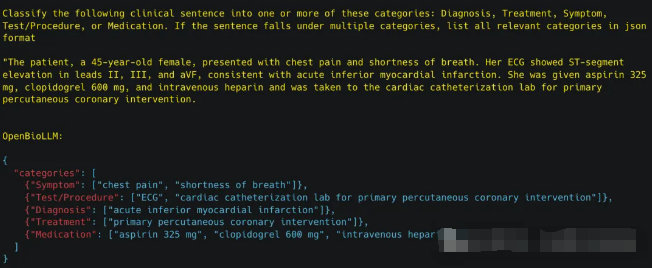
4.注意事项
虽然OpenBioLLM-70B和8B利用了高质量的数据源,但其输出仍可能包含不准确,偏差或错位,如果依赖这些不准确,偏差或错位,如果不进行进一步的测试和改进,可能会带来风险。该模型的性能尚未在随机对照试验或真实世界的医疗保健环境中进行严格评估。因此,我们强烈建议目前不要将OpenBioLLM-70B和8B用于任何直接的患者护理,临床决策支持或其他专业医疗目的。它的使用应仅限于了解其局限性的合格人员的研究、开发和探索性应用。OpenBioLLM-70B和8B仅作为协助医疗保健专业人员的研究工具,绝不应被视为合格医生的专业判断和专业知识的替代品。针对特定的医疗用例适当调整和验证OpenBioLLM-70B和8B将需要大量的额外工作,可能包括:
- 在相关临床场景中进行全面测试和评估
- 与循证指南和最佳实践保持一致
- 减轻潜在的偏差和故障模式
- 与人工监督和解释相结合
- 遵守法规和道德标准
如何学习AI大模型?
作为一名热心肠的互联网老兵,我决定把宝贵的AI知识分享给大家。 至于能学习到多少就看你的学习毅力和能力了 。我已将重要的AI大模型资料包括AI大模型入门学习思维导图、精品AI大模型学习书籍手册、视频教程、实战学习等录播视频免费分享出来。
这份完整版的大模型 AI 学习资料已经上传CSDN,朋友们如果需要可以微信扫描下方CSDN官方认证二维码免费领取【保证100%免费】

一、全套AGI大模型学习路线
AI大模型时代的学习之旅:从基础到前沿,掌握人工智能的核心技能!

二、640套AI大模型报告合集
这套包含640份报告的合集,涵盖了AI大模型的理论研究、技术实现、行业应用等多个方面。无论您是科研人员、工程师,还是对AI大模型感兴趣的爱好者,这套报告合集都将为您提供宝贵的信息和启示。

三、AI大模型经典PDF籍
随着人工智能技术的飞速发展,AI大模型已经成为了当今科技领域的一大热点。这些大型预训练模型,如GPT-3、BERT、XLNet等,以其强大的语言理解和生成能力,正在改变我们对人工智能的认识。 那以下这些PDF籍就是非常不错的学习资源。
四、AI大模型商业化落地方案

作为普通人,入局大模型时代需要持续学习和实践,不断提高自己的技能和认知水平,同时也需要有责任感和伦理意识,为人工智能的健康发展贡献力量。





















 87
87

 被折叠的 条评论
为什么被折叠?
被折叠的 条评论
为什么被折叠?








